Abstract
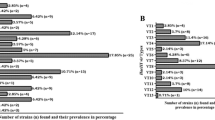
Two phenotypic and three molecular methods were assessed for their ability to identify viridans group streptococci (VGS) to the species level. A panel of 23 clinical isolates, comprising strains isolated from infective endocarditis, blood cultures, pleural and peritoneal fluid, and 19 type/reference strains were analyzed. Identification was performed using two conventional phenotypic methods: API® rapid ID 32 Strep and the VITEK® 2 system, and genotypic analysis of the nucleotide sequence of the housekeeping gene sodA, restriction patterns generated by restriction fragment length polymorphism (RFLP) of the 16S rRNA gene and multilocus sequence analysis (MLSA) of seven housekeeping genes. The API® rapid ID 32 Strep accurately speciated 79% of the strains assessed, while the VITEK® 2 generated a successful identification for 55%, presenting limitations particularly with regard to species belonging to the mitis group. RFLP of the 16S rRNA gene correctly speciated 24% of the strains, having failed to allocate a species for 36% of the isolates examined. In contrast, sequence analysis of the sodA gene provided a correct identification for 95% of the strains assessed, while identification using the MLSA technique was unsuccessful due to practical limitations. The results generated herein indicate that no single methodology can be used to provide an accurate identification to the species level of all VGS, although nucleotide sequence analysis of the sodA gene proved to be useful in providing reliable speciation.

Similar content being viewed by others
References
Alam S, Brailsford SR, Whiley RA, Beighton D (1999) PCR-Based methods for genotyping viridans group streptococci. J Clin Microbiol 37(9):2772–2776
Hakenbeck R (2007) Molecular biology of streptococci. Horizon Bioscience, Wymondham, UK
Amoroso A, Demares D, Mollerach M, Gutkind G, Coyette J (2001) All detectable high-molecular-mass penicillin-binding proteins are modified in a high-level beta-lactam-resistant clinical isolate of Streptococcus mitis. Antimicrob Agents Chemother 45(7):2075–2081
Oho T, Yamashita Y, Shimazaki Y, Kushiyama M, Koga T (2000) Simple and rapid detection of Streptococcus mutans and Streptococcus sobrinus in human saliva by polymerase chain reaction. Oral Microbiol Immunol 15(4):258–262
Beynon RP, Bahl VK, Prendergast BD (2006) Infective endocarditis. BMJ 333(7563):334–339
Chen CC, Teng LJ, Chang TC (2004) Identification of clinically relevant viridans group streptococci by sequence analysis of the 16S-23S ribosomal DNA spacer region. J Clin Microbiol 42(6):2651–2657
Claridge JE 3rd, Attorri S, Musher DM, Hebert J, Dunbar S (2001) Streptococcus intermedius, Streptococcus constellatus, and Streptococcus anginosus ("Streptococcus milleri group") are of different clinical importance and are not equally associated with abscess. Clin Infect Dis 32(10):1511–1515
Tunkel AR, Sepkowitz KA (2002) Infections caused by viridans streptococci in patients with neutropenia. Clin Infect Dis 34(11):1524–1529
Shenep JL (2000) Viridans-group streptococcal infections in immunocompromised hosts. Int J Antimicrob Agents 14(2):129–135
Boudewijns M, Bakkers JM, Sturm PD, Melchers WJ (2006) 16S rRNA gene sequencing and the routine clinical microbiology laboratory: a perfect marriage? J Clin Microbiol 44(9):3469–3470
Petti CA, Polage CR, Schreckenberger P (2005) The role of 16S rRNA gene sequencing in identification of microorganisms misidentified by conventional methods. J Clin Microbiol 43(12):6123–6125
Yuan S, Astion ML, Schapiro J, Limaye AP (2005) Clinical impact associated with corrected results in clinical microbiology testing. J Clin Microbiol 43(5):2188–2193
Haanpera M, Jalava J, Huovinen P, Meurman O, Rantakokko-Jalava K (2007) Identification of alpha-hemolytic streptococci by pyrosequencing the 16S rRNA gene and by use of VITEK 2. J Clin Microbiol 45(3):762–770
Jayarao BM, Oliver SP, Matthews KR, King SH (1991) Comparative evaluation of Vitek gram-positive identification system and API Rapid Strep system for identification of Streptococcus species of bovine origin. Vet Microbiol 26(3):301–308
Lupetti A, Barnini S, Castagna B, Capria AL, Nibbering PH (2010) Rapid identification and antimicrobial susceptibility profiling of Gram-positive cocci in blood cultures with the Vitek 2 system. Eur J Clin Microbiol Infect Dis 29(1):89–95
Innings A, Krabbe M, Ullberg M, Herrmann B (2005) Identification of 43 Streptococcus species by pyrosequencing analysis of the rnpB gene. J Clin Microbiol 43(12):5983–5991
Hoshino T, Fujiwara T, Kilian M (2005) Use of phylogenetic and phenotypic analyses to identify nonhemolytic streptococci isolated from bacteremic patients. J Clin Microbiol 43(12):6073–6085
Poyart C, Quesne G, Coulon S, Berche P, Trieu-Cuot P (1998) Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J Clin Microbiol 36(1):41–47
Baseggio N, Mansell PD, Browning JW, Browning GF (1997) Strain differentiation of isolates of streptococci from bovine mastitis by pulsed-field gel electrophoresis. Mol Cell Probes 11(5):349–354
Mineyama R, Yoshino S, Maeda N (2006) DNA fingerprinting of isolates of Streptococcus mutans by pulsed-field gel electrophoresis. Microbiol Res 162(3):244–249
Sato T, Hu JP, Ohki K, Yamaura M, Washio J, Matsuyama J, Takahashi N (2003) Identification of mutans streptococci by restriction fragment length polymorphism analysis of polymerase chain reaction-amplified 16S ribosomal RNA genes. Oral Microbiol Immunol 18(5):323–326
Jang J, Kim B, Lee J, Han H (2003) A rapid method for identification of typical Leuconostoc species by 16S rDNA PCR-RFLP analysis. J Microbiol Methods 55(1):295–302
Mirhendi H, Makimura K, Zomorodian K, Yamada T, Sugita T, Yamaguchi H (2005) A simple PCR-RFLP method for identification and differentiation of 11 Malassezia species. J Microbiol Methods 61(2):281–284
Rudney JD, Neuvar EK, Soberay AH (1992) Restriction endonuclease-fragment polymorphisms of oral viridans streptococci, compared by conventional and field-inversion gel electrophoresis. J Dent Res 71(5):1182–1188
Bishop CJ, Aanensen DM, Jordan GE, Kilian M, Hanage WP, Spratt BG (2009) Assigning strains to bacterial species via the internet. BMC Biol 7:3
Facklam R (2002) What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin Microbiol Rev 15(4):613–630
Freney J, Bland S, Etienne J, Desmonceaux M, Boeufgras JM, Fleurette J (1992) Description and evaluation of the semiautomated 4-hour rapid ID 32 Strep method for identification of streptococci and members of related genera. J Clin Microbiol 30(10):2657–2661
Jacobs JA, Schot CS, Bunschoten AE, Schouls LM (1996) Rapid species identification of "Streptococcus milleri" strains by line blot hybridization: identification of a distinct 16S rRNA population closely related to Streptococcus constellatus. J Clin Microbiol 34(7):1717–1721
Limia A, Alarcon T, Jimenez ML, Lopez-Brea M (2000) Comparison of three methods for identification of Streptococcus milleri group isolates to species level. Eur J Clin Microbiol Infect Dis 19(2):128–131
Bosshard PP, Abels S, Altwegg M, Bottger EC, Zbinden R (2004) Comparison of conventional and molecular methods for identification of aerobic catalase-negative gram-positive cocci in the clinical laboratory. J Clin Microbiol 42(5):2065–2073
Ligozzi M, Bernini C, Bonora MG, De Fatima M, Zuliani J, Fontana R (2002) Evaluation of the VITEK 2 system for identification and antimicrobial susceptibility testing of medically relevant gram-positive cocci. J Clin Microbiol 40(5):1681–1686
Brigante G, Luzzaro F, Bettaccini A, Lombardi G, Meacci F, Pini B, Stefani S, Toniolo A (2006) Use of the Phoenix automated system for identification of Streptococcus and Enterococcus spp. J Clin Microbiol 44(9):3263–3267
Poyart C, Berche P, Trieu-Cuot P (1995) Characterization of superoxide dismutase genes from gram-positive bacteria by polymerase chain reaction using degenerate primers. FEMS Microbiol Lett 131(1):41–45
Kawamura Y, Hou XG, Sultana F, Miura H, Ezaki T (1995) Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int J Syst Bacteriol 45(2):406–408
Acknowledgements
The authors would like to acknowledge Dr Simon Warwick, Department of Microbiology, Barts and The London NHS Trust, London, UK, for providing strains included in this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Teles, C., Smith, A., Ramage, G. et al. Identification of clinically relevant viridans group streptococci by phenotypic and genotypic analysis. Eur J Clin Microbiol Infect Dis 30, 243–250 (2011). https://doi.org/10.1007/s10096-010-1076-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-010-1076-y