Abstract
Background
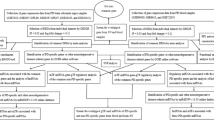
Diagnosis of Parkinson’s disease (PD) is associated with a vast number of challenges. This study aimed to assess the overlap of PD patients’ transcriptomes in the substantia nigra (SN) with peripheral blood mononuclear cells (PBMCs) to discover potential biomarkers for diagnosis.
Methods
GEO data were used to select genes with significant changes in expression level in the SN region and eligible studies. Also, transcriptome data related to blood of PD patients with other neurodegenerative diseases (ND) was considered. Differential expression genes between PD and control were evaluated in the SN and blood, and RT-qPCR was applied to validate the findings.
Results
At the expression level, no significant similarity in long non-coding RNA was found between the patients’ SN and blood. While in silico results revealed 16 common mRNAs in SN and blood with significant expression levels. Among all overexpressed mRNAs, HSPA1A/B expression level had the highest expression difference between control and PD samples. Moreover, DGKH had the highest score of down-regulated genes in both blood and SN. The NOTCH pathway had the highest score pathway among up-regulated pathways, and the expression levels of NOTCH2, H4C8, and H2BC21 associated with this pathway had the most ability to separate the control and PD populations. Furthermore, RT-qPCR results revealed that HSPA1A/B, NOTCH2, and H4C8 were overexpressed in PD PBMCs, while DGKH expression levels were lower compared to controls.
Conclusion
Our findings indicate that expression levels of HSPA1A/B, DGKH, and NOTCH2 could be applied as candidate biomarkers to diagnose PD patients in the SN region and PBMCs.




Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article.
Abbreviations
- PD:
-
Parkinson’s disease
- SN:
-
Substantia nigra
- PBMCs:
-
Peripheral blood mononuclear cells
- ND:
-
Neurodegenerative diseases
- HD:
-
Huntington’s disease
- MSA:
-
Multiple system atrophy
- PSP:
-
Progressive supranuclear palsy
- ROC:
-
Receiver operating characteristic
- GSEA:
-
Gene set enrichment analysis
References
Abdullah R, Basak I, Patil KS et al (2015) Parkinson’s disease and age: the obvious but largely unexplored link. Exp Gerontol 68:33–38. https://doi.org/10.1016/j.exger.2014.09.014
Armstrong RA, Lantos PL, Cairns NJ (2005) Overlap between neurodegenerative disorders. Neuropathology 25:111–124. https://doi.org/10.1111/j.1440-1789.2005.00605.x
Davie CA (2008) A review of Parkinson’s disease. Br Med Bull 86:109–127. https://doi.org/10.1093/bmb/ldn013
Jankovic J (2008) Parkinson’s disease: clinical features and diagnosis. J Neurol Neurosurg Psychiatry 79:368–376. https://doi.org/10.1136/jnnp.2007.131045
Chahine LM, Stern MB, Chen-Plotkin A (2014) Blood-based biomarkers for Parkinson’s disease. Park Relat Disord 20:S99–S103. https://doi.org/10.1016/S1353-8020(13)70025-7
Le W, Pan T, Huang M et al (2008) Decreased NURR1 gene expression in patients with Parkinson’s disease. J Neurol Sci 273:29–33. https://doi.org/10.1016/j.jns.2008.06.007
Yang Z, Li T, Li S et al (2019) Altered expression levels of microRNA-132 and NURR1 in peripheral blood of Parkinson’s disease: potential disease biomarkers. ACS Chem Neurosci 10:2243–2249. https://doi.org/10.1021/acschemneuro.8b00460
de Aguiar PMC, Severino P (2010) Biomarkers in Parkinson disease: global gene expression analysis in peripheral blood from patients with and without mutations in PARK2 and PARK8. Einstein (São Paulo) 8:291–297. https://doi.org/10.1590/s1679-45082010ao1674
Fazeli S, Motovali-Bashi M, Peymani M et al (2020) A compound downregulation of SRRM2 and miR-27a-3p with upregulation of miR-27b-3p in PBMCs of Parkinson’s patients is associated with the early stage onset of disease. PLoS One 15:e0240855. https://doi.org/10.1371/journal.pone.0240855
Smyth GK (2005) limma: linear models for microarray data. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer-Verlag, New York, pp 397–420
Leek JT, Johnson WE, Parker HS et al (2012) The SVA package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28:882–883. https://doi.org/10.1093/bioinformatics/bts034
Fahn S, Marsden CD, Calne DB, Goldstein M (1987) Recent developments in Parkinson’s disease, vol 2. Florham Park, NJ Macmillan Healthc Inf, p 15
Elbaz A, Carcaillon L, Kab S, Moisan F (2015) Epidemiology of Parkinson’s disease: the Rotterdam Study. Rev Neurol (Paris) 1580:1–13. https://doi.org/10.1016/j.neurol.2015.09.012
Minoia M, Grit C, Kampinga HH (2014) HSPA1A-independent suppression of PARK2 C289G protein aggregation by human small heat shock proteins. Mol Cell Biol 34:3570–3578. https://doi.org/10.1128/mcb.00698-14
Ekimova IV, Pazi MB, Belan DV (2019) The impact of pharmacological inhibition of HSP70 chaperone expression on protective effects of the glucose-regulated 78 kDa protein in a Parkinson’s disease model. J Evol Biochem Physiol 55:419–422. https://doi.org/10.1134/S0022093019050107
Bossers K, Meerhoff G, Balesar R et al (2009) Analysis of gene expression in Parkinson’s disease: possible involvement of neurotrophic support and axon guidance in dopaminergic cell death. Brain Pathol 19:91–107. https://doi.org/10.1111/j.1750-3639.2008.00171.x
Sakane F, Mizuno S, Komenoi S (2016) Diacylglycerol kinases as emerging potential drug targets for a variety of diseases: an update. Front Cell Dev Biol 4. https://doi.org/10.3389/fcell.2016.00082
Wood PL, Tippireddy S, Feriante J, Woltjer RL (2018) Augmented frontal cortex diacylglycerol levels in Parkinson’s disease and Lewy body disease. PLoS One 13:e0191815. https://doi.org/10.1371/journal.pone.0191815
Imai Y, Kobayashi Y, Inoshita T et al (2015) The Parkinson’s disease-associated protein kinase LRRK2 modulates Notch signaling through the endosomal pathway. PLoS Genet 11:e1005503. https://doi.org/10.1371/journal.pgen.1005503
Dong M-X, Feng X, Xu X-M et al (2018) Integrated analysis reveals altered lipid and glucose metabolism and identifies NOTCH2 as a biomarker for Parkinson’s disease related depression. Front Mol Neurosci 11. https://doi.org/10.3389/fnmol.2018.00257
Park G, Tan J, Garcia G et al (2016) Regulation of histone acetylation by autophagy in Parkinson disease. J Biol Chem 291:3531–3540. https://doi.org/10.1074/jbc.M115.675488
Acknowledgements
We thank Mr. Mohammad Mahdevar for his association in RNA-seq and microarray analysis. Also, we thank our colleagues for their helpful discussions in this study. This research was partly supported by TIG Grant Committee (Avicenna Center of excellence) and Isfahan Research Network (project title: Neurodegenerative Diseases Research Group) to K.G.
Author information
Authors and Affiliations
Contributions
The design, sample collection, and conceptualization of study and methodology were done by Leila Asad Samani and Maryam Peymani. Data mining, formal analysis, and investigation were performed by Leila Asad Samani. Supervision, validation, and visualization were done by Maryam Peymani and Kamran Ghaedi. Interpretation of the obtained information was done by Maryam Peymani and sample collection by Masoud Etemadifar. The manuscript was written by Leila Asad Samani. Review, editing, and approval were by Maryam Peymani, Kamran Ghaedi, and Ahmad Majd. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval
All protocols for the usage of human samples in this study were reviewed and approved by the Biomedical Ethics Committee of the Islamic Azad University—North Tehran Branch and also complies with the Ethics Code of IR.IAU.TNB.REC.1400.007 in accordance with the relevant guidelines and regulations for using of human samples.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Asad Samani, L., Ghaedi, K., Majd, A. et al. Coordinated modification in expression levels of HSPA1A/B, DGKH, and NOTCH2 in Parkinson’s patients’ blood and substantia nigra as a diagnostic sign: the transcriptomes’ relationship. Neurol Sci 44, 2753–2761 (2023). https://doi.org/10.1007/s10072-023-06738-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10072-023-06738-4