Abstract
Objectives
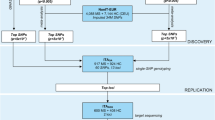
To validate in an ethnically homogeneous Greek multiple sclerosis (MS) cohort, genetic risk factors for the disease, identified through a number of previous multi-ethnic genome-wide association studies (GWAS).
Methods
A total of 1228 MS cases and 1014 controls were recruited in the study, from 3 MS centers in Greece. We genotyped 35 susceptibility SNPs that emerged from previous GWAS or meta-analyses of GWAS. Allele and genotype single locus regression analysis, adjusted for gender and site, was performed. Permutation testing was applied to all analyses.
Results
Six polymorphisms reached statistical significance (permutation p value < 0.05). In particular, rs2760524 of LOC105371664, near RGS1 (permutation p value 0.001), rs3129889 of HLA-DRA, near HLA-DRB1 (permutation p value < 1.00e-04), rs1738074 of TAGAP (permutation p value 0.007), rs703842 of METTL1/CYP27B1 (permutation p value 0.008), rs9596270 of DLEU1 (permutation p value < 1.00e-04), and rs17445836 of LincRNA, near IRF8 (permutation p value 0.001) were identified as susceptibility risk factors in our group.
Conclusion
The current study replicated a number of GWAS susceptibility SNPs, which implies that some similarities between the examined Greek population and the MS genetic architecture of the GWAS populations do exist.
Similar content being viewed by others
References
Compston A, Coles A (2008) Multiple sclerosis. Lancet (London, England) 372(9648):1502–1517. https://doi.org/10.1016/s0140-6736(08)61620-7
Compston A, Sadovnick AD (1992) Epidemiology and genetics of multiple sclerosis. Curr Opin Neurol Neurosurg 5(2):175–181
Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20 (2009) Nat Genet 41(7):824–828. https://doi.org/10.1038/ng.396
Sokratous M, Dardiotis E, Bellou E, Tsouris Z, Michalopoulou A, Dardioti M, Siokas V, Rikos D, Tsatsakis A, Kovatsi L, Bogdanos DP, Hadjigeorgiou GM (2018) CpG Island methylation patterns in relapsing-remitting multiple sclerosis. J Mol Neurosci: MN 64(3):478–484. https://doi.org/10.1007/s12031-018-1046-x
Sokratous M, Dardiotis E, Tsouris Z, Bellou E, Michalopoulou A, Siokas V, Arseniou S, Stamati T, Tsivgoulis G, Bogdanos D, Hadjigeorgiou GM (2016) Deciphering the role of DNA methylation in multiple sclerosis: emerging issues. Auto- immunity highlights 7(1):12. https://doi.org/10.1007/s13317-016-0084-z
Bomprezzi R, Kovanen PE, Martin R (2003) New approaches to investigating heterogeneity in complex traits. J Med Genet 40(8):553–559
Mentis AA, Dardiotis E, Grigoriadis N, Petinaki E, Hadjigeorgiou GM (2017) Viruses and multiple sclerosis: from mechanisms and pathways to translational research opportunities. Mol Neurobiol 54(5):3911–3923. https://doi.org/10.1007/s12035-017-0530-6
Trapp BD, Vignos M, Dudman J, Chang A, Fisher E, Staugaitis SM, Battapady H, Mork S, Ontaneda D, Jones SE, Fox RJ, Chen J, Nakamura K, Rudick RA (2018) Cortical neuronal densities and cerebral white matter demyelination in multiple sclerosis: a retrospective study. Lancet Neurol. https://doi.org/10.1016/S1474-4422(18)30245-X
Zarei M, Chandran S, Compston A, Hodges J (2003) Cognitive presentation of multiple sclerosis: evidence for a cortical variant. J Neurol Neurosurg Psychiatry 74(7):872–877
Bashinskaya VV, Kulakova OG, Boyko AN, Favorov AV, Favorova OO (2015) A review of genome-wide association studies for multiple sclerosis: classical and hypothesis-driven approaches. Hum Genet 134(11–12):1143–1162. https://doi.org/10.1007/s00439-015-1601-2
Kowalec K, Wright GEB, Drogemoller BI, Aminkeng F, Bhavsar AP, Kingwell E, Yoshida EM, Traboulsee A, Marrie RA, Kremenchutzky M, Campbell TL, Duquette P, Chalasani N, Wadelius M, Hallberg P, Xia Z, De Jager PL, Denny JC, Davis MF, Ross CJD, Tremlett H, Carleton BC (2018) Common variation near IRF6 is associated with IFN-beta-induced liver injury in multiple sclerosis. Nat Genet 50(8):1081–1085. https://doi.org/10.1038/s41588-018-0168-y
Dardiotis E, Panayiotou E, Provatas A, Christodoulou K, Hadjisavvas A, Antoniades A, Lourbopoulos A, Pantzaris M, Grigoriadis N, Hadjigeorgiou GM, Kyriakides T (2017) Gene variants of adhesion molecules act as modifiers of disease severity in MS. Neurology(R) Neuroimmunol Neuroinflammation 4(4):e350. https://doi.org/10.1212/nxi.0000000000000350
Sawcer S, Franklin RJ, Ban M (2014) Multiple sclerosis genetics. Lancet Neurol 13(7):700–709. https://doi.org/10.1016/s1474-4422(14)70041-9
Axisa PP, Hafler DA (2016) Multiple sclerosis: genetics, biomarkers, treatments. Curr Opin Neurol 29(3):345–353. https://doi.org/10.1097/wco.0000000000000319
Lill CM (2014) Recent advances and future challenges in the genetics of multiple sclerosis. Front Neurol 5:130. https://doi.org/10.3389/fneur.2014.00130
Torkamani A, Pham P, Libiger O, Bansal V, Zhang G, Scott-Van Zeeland AA, Tewhey R, Topol EJ, Schork NJ (2012) Clinical implications of human population differences in genome-wide rates of functional genotypes. Front Genet 3:211. https://doi.org/10.3389/fgene.2012.00211
Patsopoulos NA, Bayer Pharma MSGWG, Steering Committees of Studies Evaluating I-b, a CCRA, Consortium AN, GeneMsa, International Multiple Sclerosis Genetics C, Esposito F, Reischl J, Lehr S, Bauer D, Heubach J, Sandbrink R, Pohl C, Edan G, Kappos L, Miller D, Montalban J, Polman CH, Freedman MS, Hartung HP, Arnason BG, Comi G, Cook S, Filippi M, Goodin DS, Jeffery D, O'Connor P, Ebers GC, Langdon D, Reder AT, Traboulsee A, Zipp F, Schimrigk S, Hillert J, Bahlo M, Booth DR, Broadley S, Brown MA, Browning BL, Browning SR, Butzkueven H, Carroll WM, Chapman C, Foote SJ, Griffiths L, Kermode AG, Kilpatrick TJ, Lechner-Scott J, Marriott M, Mason D, Moscato P, Heard RN, Pender MP, Perreau VM, Perera D, Rubio JP, Scott RJ, Slee M, Stankovich J, Stewart GJ, Taylor BV, Tubridy N, Willoughby E, Wiley J, Matthews P, Boneschi FM, Compston A, Haines J, Hauser SL, McCauley J, Ivinson A, Oksenberg JR, Pericak-Vance M, Sawcer SJ, De Jager PL, Hafler DA, de Bakker PI (2011) Genome-wide meta-analysis identifies novel multiple sclerosis susceptibility loci. Ann Neurol 70(6):897–912. https://doi.org/10.1002/ana.22609
Aggelakis K, Zacharaki F, Dardiotis E, Xiromerisiou G, Tsimourtou V, Ralli S, Gkaraveli M, Bourpoulas D, Rodopoulou P, Papadimitriou A, Hadjigeorgiou G (2010) Interleukin-1B and interleukin-1 receptor antagonist gene polymorphisms in Greek multiple sclerosis (MS) patients with bout-onset MS. Neurol Sci 31(3):253–257. https://doi.org/10.1007/s10072-009-0155-2
Bauchet M, McEvoy B, Pearson LN, Quillen EE, Sarkisian T, Hovhannesyan K, Deka R, Bradley DG, Shriver MD (2007) Measuring European population stratification with microarray genotype data. Am J Hum Genet 80(5):948–956. https://doi.org/10.1086/513477
Shi YY, He L (2005) SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res 15(2):97–98. https://doi.org/10.1038/sj.cr.7290272
Spencer CC, Su Z, Donnelly P, Marchini J (2009) Designing genome-wide association studies: sample size, power, imputation, and the choice of genotyping chip. PLoS Genet 5(5):e1000477. https://doi.org/10.1371/journal.pgen.1000477
Soundararajan M, Willard FS, Kimple AJ, Turnbull AP, Ball LJ, Schoch GA, Gileadi C, Fedorov OY, Dowler EF, Higman VA, Hutsell SQ, Sundstrom M, Doyle DA, Siderovski DP (2008) Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits. Proc Natl Acad Sci U S A 105(17):6457–6462. https://doi.org/10.1073/pnas.0801508105
The International Multiple Sclerosis Genetics C (2010) IL12A, MPHOSPH9/CDK2AP1 and RGS1 are novel multiple sclerosis susceptibility loci. Genes Immun 11(5):397–405. https://doi.org/10.1038/gene.2010.28
Tran T, Paz P, Velichko S, Cifrese J, Belur P, Yamaguchi KD, Ku K, Mirshahpanah P, Reder AT, Croze E (2010) Interferonbeta-1b induces the expression of RGS1 a negative regulator of G-protein signaling. Int J Cell Biol 2010:529376. https://doi.org/10.1155/2010/529376
De Jager PL, Jia X, Wang J, de Bakker PI, Ottoboni L, Aggarwal NT, Piccio L, Raychaudhuri S, Tran D, Aubin C, Briskin R, Romano S, Baranzini SE, McCauley JL, Pericak-Vance MA, Haines JL, Gibson RA, Naeglin Y, Uitdehaag B, Matthews PM, Kappos L, Polman C, McArdle WL, Strachan DP, Evans D, Cross AH, Daly MJ, Compston A, Sawcer SJ, Weiner HL, Hauser SL, Hafler DA, Oksenberg JR (2009) Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet 41(7):776–782. https://doi.org/10.1038/ng.401
Jiang T, Li L, Wang Y, Zhao C, Yang J, Ma D, Guan Y, Zhao D, Bao Y, Wang Y, Yang J (2016) The association between genetic polymorphism rs703842 in CYP27B1 and multiple sclerosis: a meta-analysis. Medicine 95(19):e3612. https://doi.org/10.1097/md.0000000000003612
Jones G, Prosser DE, Kaufmann M (2014) Cytochrome P450-mediated metabolism of vitamin D. J Lipid Res 55(1):13–31. https://doi.org/10.1194/jlr.R031534
Alharbi FM (2015) Update in vitamin D and multiple sclerosis. Neurosciences 20(4):329–335. https://doi.org/10.17712/nsj.2015.4.20150357
Ramagopalan SV, Dyment DA, Cader MZ, Morrison KM, Disanto G, Morahan JM, Berlanga-Taylor AJ, Handel A, De Luca GC, Sadovnick AD, Lepage P, Montpetit A, Ebers GC (2011) Rare variants in the CYP27B1 gene are associated with multiple sclerosis. Ann Neurol 70(6):881–886. https://doi.org/10.1002/ana.22678
Runia TF, Hop WC, de Rijke YB, Buljevac D, Hintzen RQ (2012) Lower serum vitamin D levels are associated with a higher relapse risk in multiple sclerosis. Neurology 79(3):261–266. https://doi.org/10.1212/WNL.0b013e31825fdec7
Berge T, Leikfoss IS, Brorson IS, Bos SD, Page CM, Gustavsen MW, Bjolgerud A, Holmoy T, Celius EG, Damoiseaux J, Smolders J, Harbo HF, Spurkland A (2016) The multiple sclerosis susceptibility genes TAGAP and IL2RA are regulated by vitamin D in CD4+ T cells. Genes Immun 17(2):118–127. https://doi.org/10.1038/gene.2015.61
Huang SQ, Zhang N, Zhou ZX, Huang CC, Zeng CL, Xiao D, Guo CC, Han YJ, Ye XH, Ye XG, Ou ML, Zhang BH, Liu Y, Zeng EY, Yang G, Jing CX (2017) Association of LPP and TAGAP polymorphisms with celiac disease risk: a meta-analysis. Int J Environ Res Public Health 14(2). https://doi.org/10.3390/ijerph14020171
Beecham AH, Patsopoulos NA, Xifara DK, Davis MF, Kemppinen A, Cotsapas C, Shah TS, Spencer C, Booth D, Goris A, Oturai A, Saarela J, Fontaine B, Hemmer B, Martin C, Zipp F, D'Alfonso S, Martinelli-Boneschi F, Taylor B, Harbo HF, Kockum I, Hillert J, Olsson T, Ban M, Oksenberg JR, Hintzen R, Barcellos LF, Agliardi C, Alfredsson L, Alizadeh M, Anderson C, Andrews R, Sondergaard HB, Baker A, Band G, Baranzini SE, Barizzone N, Barrett J, Bellenguez C, Bergamaschi L, Bernardinelli L, Berthele A, Biberacher V, Binder TM, Blackburn H, Bomfim IL, Brambilla P, Broadley S, Brochet B, Brundin L, Buck D, Butzkueven H, Caillier SJ, Camu W, Carpentier W, Cavalla P, Celius EG, Coman I, Comi G, Corrado L, Cosemans L, Cournu-Rebeix I, Cree BA, Cusi D, Damotte V, Defer G, Delgado SR, Deloukas P, di Sapio A, Dilthey AT, Donnelly P, Dubois B, Duddy M, Edkins S, Elovaara I, Esposito F, Evangelou N, Fiddes B, Field J, Franke A, Freeman C, Frohlich IY, Galimberti D, Gieger C, Gourraud PA, Graetz C, Graham A, Grummel V, Guaschino C, Hadjixenofontos A, Hakonarson H, Halfpenny C, Hall G, Hall P, Hamsten A, Harley J, Harrower T, Hawkins C, Hellenthal G, Hillier C, Hobart J, Hoshi M, Hunt SE, Jagodic M, Jelcic I, Jochim A, Kendall B, Kermode A, Kilpatrick T, Koivisto K, Konidari I, Korn T, Kronsbein H, Langford C, Larsson M, Lathrop M, Lebrun-Frenay C, Lechner-Scott J, Lee MH, Leone MA, Leppa V, Liberatore G, Lie BA, Lill CM, Linden M, Link J, Luessi F, Lycke J, Macciardi F, Mannisto S, Manrique CP, Martin R, Martinelli V, Mason D, Mazibrada G, McCabe C, Mero IL, Mescheriakova J, Moutsianas L, Myhr KM, Nagels G, Nicholas R, Nilsson P, Piehl F, Pirinen M, Price SE, Quach H, Reunanen M, Robberecht W, Robertson NP, Rodegher M, Rog D, Salvetti M, Schnetz-Boutaud NC, Sellebjerg F, Selter RC, Schaefer C, Shaunak S, Shen L, Shields S, Siffrin V, Slee M, Sorensen PS, Sorosina M, Sospedra M, Spurkland A, Strange A, Sundqvist E, Thijs V, Thorpe J, Ticca A, Tienari P, van Duijn C, Visser EM, Vucic S, Westerlind H, Wiley JS, Wilkins A, Wilson JF, Winkelmann J, Zajicek J, Zindler E, Haines JL, Pericak-Vance MA, Ivinson AJ, Stewart G, Hafler D, Hauser SL, Compston A, McVean G, De Jager P, Sawcer SJ, McCauley JL (2013) Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet 45(11):1353–1360. https://doi.org/10.1038/ng.2770
Mc Guire C, Prinz M, Beyaert R, van Loo G (2013) Nuclear factor kappa B (NF-kappaB) in multiple sclerosis pathology. Trends Mol Med 19(10):604–613. https://doi.org/10.1016/j.molmed.2013.08.001
Leibowitz SM, Yan J (2016) NF-κB pathways in the pathogenesis of multiple sclerosis and the therapeutic implications. Front Mol Neurosci 9:84. https://doi.org/10.3389/fnmol.2016.00084
Peng H, Guerau-de-Arellano M, Mehta VB, Yang Y, Huss DJ, Papenfuss TL, Lovett-Racke AE, Racke MK (2012) Dimethyl fumarate inhibits dendritic cell maturation via nuclear factor kappaB (NF-kappaB) and extracellular signal-regulated kinase 1 and 2 (ERK1/2) and mitogen stress-activated kinase 1 (MSK1) signaling. J Biol Chem 287(33):28017–28026. https://doi.org/10.1074/jbc.M112.383380
Dyment DA, Herrera BM, Cader MZ, Willer CJ, Lincoln MR, Sadovnick AD, Risch N, Ebers GC (2005) Complex interactions among MHC haplotypes in multiple sclerosis: susceptibility and resistance. Hum Mol Genet 14(14):2019–2026. https://doi.org/10.1093/hmg/ddi206
Kouri I, Papakonstantinou S, Bempes V, Vasiliadis HS, Kyritsis AP, Pelidou SH (2011) HLA associations with multiple sclerosis in Greece. J Neurol Sci 308(1–2):28–31. https://doi.org/10.1016/j.jns.2011.06.037
Dardiotis E, Arseniou S, Sokratous M, Tsouris Z, Siokas V, Mentis AA, Michalopoulou A, Andravizou A, Dastamani M, Paterakis K, Bogdanos D, Brotis A (2017) Vitamin B12, folate, and homocysteine levels and multiple sclerosis: a meta-analysis. Multiple sclerosis and related disorders 17:190–197. https://doi.org/10.1016/j.msard.2017.08.004
Mentis AA, Dardiotis E, Grigoriadis N, Petinaki E, Hadjigeorgiou GM (2017) Viruses and endogenous retroviruses in multiple sclerosis: from correlation to causation. Acta Neurol Scand 136(6):606–616. https://doi.org/10.1111/ane.12775
Funding
The study was supported in part by a research grant of the Research Committee of the University of Thessaly, Greece (Code: 2845).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.
Electronic supplementary material
ESM 1
(DOCX 29 kb)
Rights and permissions
About this article
Cite this article
Hadjigeorgiou, G.M., Kountra, PM., Koutsis, G. et al. Replication study of GWAS risk loci in Greek multiple sclerosis patients. Neurol Sci 40, 253–260 (2019). https://doi.org/10.1007/s10072-018-3617-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10072-018-3617-6