Abstract
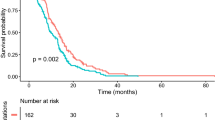
This study aimed to investigate the impact of TP53 alteration on survival and clinicopathological features of glioma patients with H3K27M mutations. An individual-participant-data (IPD) meta-analysis was performed to investigate the impact of TP53 alteration on survival and clinicopathological features of patients with H3K27M mutations. Three hundred thirty-one individual records from 12 eligible glioma studies involving the H3K27M mutation were finally included in our meta-analysis, and a pooled hazard ratio (HR) of 1.53 (95%CI, 1.10–2.11; P = 0.01) indicated that TP53 alterations were associated with a shorter overall survival. The pooled odds ratios (ORs) indicated that TP53 alterations were significantly associated with the age at diagnosis ≥ 7 years (OR = 1.97, 95%CI = 1.15–3.38, P = 0.01), the status of histone H3.3 mutations (OR = 9.15, 95%CI = 4.18–20.06, P < 0.00001), and high WHO grade histology (III + IV) (OR = 2.70, 95%CI = 1.33–5.48, P = 0.006). However, no association was found between TP53 alterations and gender or tumor location. This IPD meta-analysis suggests that TP53 alteration is a valuable predictor for the prognosis of patients with H3K27M mutated gliomas. TP53 alteration may be used for identifying a subset of patients who potentially benefit from targeted reactivation of TP53 activity.




Similar content being viewed by others
References
Schwartzentruber J, Korshunov A, Liu XY, Jones DTW, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DAK, Tönjes M, Hovestadt V, Albrecht S, Kool M, Nantel A, Konermann C, Lindroth A, Jäger N, Rausch T, Ryzhova M, Korbel JO, Hielscher T, Hauser P, Garami M, Klekner A, Bognar L, Ebinger M, Schuhmann MU, Scheurlen W, Pekrun A, Frühwald MC, Roggendorf W, Kramm C, Dürken M, Atkinson J, Lepage P, Montpetit A, Zakrzewska M, Zakrzewski K, Liberski PP, Dong Z, Siegel P, Kulozik AE, Zapatka M, Guha A, Malkin D, Felsberg J, Reifenberger G, von Deimling A, Ichimura K, Collins VP, Witt H, Milde T, Witt O, Zhang C, Castelo-Branco P, Lichter P, Faury D, Tabori U, Plass C, Majewski J, Pfister SM, Jabado N (2012) Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482:226–231
Wu G, Broniscer A, McEachron TA et al (2012) Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet 44:251–253
Zhang RQ, Shi Z, Chen H et al (2015) Biomarker-based prognostic stratification of young adult glioblastoma. Oncotarget 7:5030–5041
Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DTW, Konermann C, Pfaff E, Tönjes M, Sill M, Bender S, Kool M, Zapatka M, Becker N, Zucknick M, Hielscher T, Liu XY, Fontebasso AM, Ryzhova M, Albrecht S, Jacob K, Wolter M, Ebinger M, Schuhmann MU, van Meter T, Frühwald MC, Hauch H, Pekrun A, Radlwimmer B, Niehues T, von Komorowski G, Dürken M, Kulozik AE, Madden J, Donson A, Foreman NK, Drissi R, Fouladi M, Scheurlen W, von Deimling A, Monoranu C, Roggendorf W, Herold-Mende C, Unterberg A, Kramm CM, Felsberg J, Hartmann C, Wiestler B, Wick W, Milde T, Witt O, Lindroth AM, Schwartzentruber J, Faury D, Fleming A, Zakrzewska M, Liberski PP, Zakrzewski K, Hauser P, Garami M, Klekner A, Bognar L, Morrissy S, Cavalli F, Taylor MD, van Sluis P, Koster J, Versteeg R, Volckmann R, Mikkelsen T, Aldape K, Reifenberger G, Collins VP, Majewski J, Korshunov A, Lichter P, Plass C, Jabado N, Pfister SM (2012) Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22:425–437
Korshunov A, Ryzhova M, Hovestadt V, Bender S, Sturm D, Capper D, Meyer J, Schrimpf D, Kool M, Northcott PA, Zheludkova O, Milde T, Witt O, Kulozik AE, Reifenberger G, Jabado N, Perry A, Lichter P, von Deimling A, Pfister SM, Jones DTW (2015) Integrated analysis of pediatric glioblastoma reveals a subset of biologically favorable tumors with associated molecular prognostic markers. Acta Neuropathol 129:669–678
Khuong-Quang DA, Buczkowicz P, Rakopoulos P, Liu XY, Fontebasso AM, Bouffet E, Bartels U, Albrecht S, Schwartzentruber J, Letourneau L, Bourgey M, Bourque G, Montpetit A, Bourret G, Lepage P, Fleming A, Lichter P, Kool M, von Deimling A, Sturm D, Korshunov A, Faury D, Jones DT, Majewski J, Pfister SM, Jabado N, Hawkins C (2012) K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol 124:439–447
Buczkowicz P, Bartels U, Bouffet E, Becher O, Hawkins C (2014) Histopathological spectrum of paediatric diffuse intrinsic pontine glioma: diagnostic and therapeutic implications. Acta Neuropathol 128:573–581
Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW (2016) The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 131:803–820
Bender S, Tang Y, Lindroth AM, Hovestadt V, Jones DTW, Kool M, Zapatka M, Northcott PA, Sturm D, Wang W, Radlwimmer B, Højfeldt JW, Truffaux N, Castel D, Schubert S, Ryzhova M, Şeker-Cin H, Gronych J, Johann PD, Stark S, Meyer J, Milde T, Schuhmann M, Ebinger M, Monoranu CM, Ponnuswami A, Chen S, Jones C, Witt O, Collins VP, von Deimling A, Jabado N, Puget S, Grill J, Helin K, Korshunov A, Lichter P, Monje M, Plass C, Cho YJ, Pfister SM (2013) Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell 24:660–672
Chan KM, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, Gupta N, Mueller S, James CD, Jenkins R, Sarkaria J, Zhang Z (2013) The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev 27:985–990
Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD (2013) Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma-K27M. Science 340:857–861
Venneti S, Garimella MT, Sullivan LM, Martinez D, Huse JT, Heguy A, Santi M, Thompson CB, Judkins AR (2013) Evaluation of histone 3 lysine 27 trimethylation (H3K27me3) and enhancer of zest 2 (EZH2) in pediatric glial and glioneuronal tumors shows decreased H3K27me3 in H3F3A K27M mutant glioblastomas. Brain Pathol 23:558–564
Gessi M, Gielen GH, Dreschmann V, Waha A, Pietsch T (2015) High frequency of H3F3A K27M mutations characterizes pediatric and adult high-grade gliomas of the spinal cord. Acta Neuropathol 130:435–437
Castel D, Philippe C, Calmon R, le Dret L, Truffaux N, Boddaert N, Pagès M, Taylor KR, Saulnier P, Lacroix L, Mackay A, Jones C, Sainte-Rose C, Blauwblomme T, Andreiuolo F, Puget S, Grill J, Varlet P, Debily MA (2015) Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol 130:815–827
Buczkowicz P, Hoeman C, Rakopoulos P, Pajovic S, Letourneau L, Dzamba M, Morrison A, Lewis P, Bouffet E, Bartels U, Zuccaro J, Agnihotri S, Ryall S, Barszczyk M, Chornenkyy Y, Bourgey M, Bourque G, Montpetit A, Cordero F, Castelo-Branco P, Mangerel J, Tabori U, Ho KC, Huang A, Taylor KR, Mackay A, Bendel AE, Nazarian J, Fangusaro JR, Karajannis MA, Zagzag D, Foreman NK, Donson A, Hegert JV, Smith A, Chan J, Lafay-Cousin L, Dunn S, Hukin J, Dunham C, Scheinemann K, Michaud J, Zelcer S, Ramsay D, Cain J, Brennan C, Souweidane MM, Jones C, Allis CD, Brudno M, Becher O, Hawkins C (2014) Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nat Genet 46:451–456
Fontebasso AM, Papillon-Cavanagh S, Schwartzentruber J, Nikbakht H, Gerges N, Fiset PO, Bechet D, Faury D, de Jay N, Ramkissoon LA, Corcoran A, Jones DTW, Sturm D, Johann P, Tomita T, Goldman S, Nagib M, Bendel A, Goumnerova L, Bowers DC, Leonard JR, Rubin JB, Alden T, Browd S, Geyer JR, Leary S, Jallo G, Cohen K, Gupta N, Prados MD, Carret AS, Ellezam B, Crevier L, Klekner A, Bognar L, Hauser P, Garami M, Myseros J, Dong Z, Siegel PM, Malkin H, Ligon AH, Albrecht S, Pfister SM, Ligon KL, Majewski J, Jabado N, Kieran MW (2014) Recurrent somatic mutations in ACVR1 in pediatric midline high-grade astrocytoma. Nat Genet 46:462–466
Taylor KR, Mackay A, Truffaux N, Butterfield YS, Morozova O, Philippe C, Castel D, Grasso CS, Vinci M, Carvalho D, Carcaboso AM, de Torres C, Cruz O, Mora J, Entz-Werle N, Ingram WJ, Monje M, Hargrave D, Bullock AN, Puget S, Yip S, Jones C, Grill J (2014) Recurrent activating ACVR1 mutations in diffuse intrinsic pontine glioma. Nat Genet 46:457–461
Wu G, Diaz AK, Paugh BS et al (2014) The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet 46:444–450
Freed-Pastor WA, Prives C (2012) Mutant p53: one name, many proteins. Genes Dev 26:1268–1286
Farnebo M, Bykov VJ, Wiman KG (2010) The p53 tumor suppressor: a master regulator of diverse cellular processes and therapeutic target in cancer. Biochem Biophys Res Commun 396:85–89
Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, vanTuinen P, Ledbetter DH, Barker DF, Nakamura Y, White R, Vogelstein B (1989) Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science 244:217–221
Finlay CA Hinds PW, Levine AJ (1989) The p53 proto-oncogene can act as a suppressor of transformation. Cell 57:1083–1093
Levine AJ, Finlay CA, Hinds PW (2004) P53 is a tumor suppressor gene. Cell 116:S67–S69
Leroy B, Fournier JL, Ishioka C, Monti P, Inga A, Fronza G, Soussi T (2013) The TP53 website: an integrative resource centre for the TP53 mutation database and TP53 mutant analysis. Nucleic Acids Res 41:D962–D969
Muller PA, Vousden KH (2014) Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 25:304–317
Ohgaki H, Kleihues P (2007) Genetic pathway to primary and secondary glioblastoma. Am J Pathol 170:1445–1453
Wang X, Chen JX, Liu JP, You C, Liu YH, Mao Q (2014) Gain of function of mutant TP53 in glioblastoma: prognosis and response to temozolomide. Ann Surg Oncol 21:1337–1344
Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327:557–560
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7:177–188
Begg CB, Mazumdar M (1994) Operating characteristics of a rank correlation test for publication bias. Biometrics 50:1088–1101
Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315:629–634
Aihara K, Mukasa A, Gotoh K et al (2013) H3F3A K27M mutations in thalamic gliomas from young adult patients. Neuro-Oncology 16:140–146
Hoffman LM, DeWire M, Ryall S et al (2016) Spatial genomic heterogeneity in diffuse intrinsic pontine and midline high-grade glioma: implications for diagnostic biopsy and targeted therapeutics. ANC 4:1
Feng J, Hao S, Pan C, Wang Y, Wu Z, Zhang J, Yan H, Zhang L, Wan H (2015) The H3.3 K27M mutation results in a poorer prognosis in brainstem gliomas than thalamic gliomas in adults. Hum Pathol 46:1626–1632
Misuraca KL, Barton KL, Chung A et al (2014) Pax3 expression enhances PDGF-B-induced brainstem gliomagenesis and characterizes a subset of brainstem glioma. ANC 2:134
Nikbakht H, Panditharatna E, Mikael LG, Li R, Gayden T, Osmond M, Ho CY, Kambhampati M, Hwang EI, Faury D, Siu A, Papillon-Cavanagh S, Bechet D, Ligon KL, Ellezam B, Ingram WJ, Stinson C, Moore AS, Warren KE, Karamchandani J, Packer RJ, Jabado N, Majewski J, Nazarian J (2016) Spatial and temporal homogeneity of driver mutations in diffuse intrinsic pontine glioma. Nat Commun 7:11185
Zhang L, Chen LH, Wan H, Yang R, Wang Z, Feng J, Yang S, Jones S, Wang S, Zhou W, Zhu H, Killela PJ, Zhang J, Wu Z, Li G, Hao S, Wang Y, Webb JB, Friedman HS, Friedman AH, McLendon RE, He Y, Reitman ZJ, Bigner DD, Yan H (2014) Exome sequencing identifies somatic gain-of-function PPM1D mutations in brainstem gliomas. Nat Genet 46:726–730
Bogler O, Huang HJ, Kleihues P, Cavenee WK (1995) The p53 gene and its role in human brain tumors. Glia 15:308–327
Migliavacca M, Ottini L, Bazan V, Agnese V, Corsale S, Macaluso M, Lupi R, Dardanoni G, Valerio MR, Pantuso G, Fede GD, Tomasino RM, Gebbia N, Mariani-Costantini R, Russo A (2004) TP53 in gastric cancer: mutations in the l3 loop and LSH motif DNA-binding domains of TP53 predict poor outcome. J Cell Physiol 200:476–485
Langerod A, Zhao H, Borgan O et al (2007) TP53 mutation status and gene expression profiles are powerful prognostic markers of breast cancer. Breast Cancer Res 9:R30
Hof J, Krentz S, van Schewick C, Körner G, Shalapour S, Rhein P, Karawajew L, Ludwig WD, Seeger K, Henze G, von Stackelberg A, Hagemeier C, Eckert C, Kirschner-Schwabe R (2011) Mutations and deletions of the TP53 gene predict nonresponse to treatment and poor outcome in first relapse of childhood acute lymphoblastic leukemia. J Clin Oncol 29:3185–3193
Funato K, Major T, Lewis PW, Allis CD, Tabar V (2014) Use of human embryonic stem cells to model pediatric gliomas with H3.3K27M histone mutation. Science 346:1529–1533
Goldberg AD, Banaszynski LA, Noh K-M, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, Wen D, Chapgier A, DeKelver RC, Miller JC, Lee YL, Boydston EA, Holmes MC, Gregory PD, Greally JM, Rafii S, Yang C, Scambler PJ, Garrick D, Gibbons RJ, Higgs DR, Cristea IM, Urnov FD, Zheng D, Allis CD (2010) Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140:678–691
Szenker E, Ray-Gallet D, Almouzni G (2011) The double face of the histone variant H3.3. Cell Res 21:421–434
Michotte A, Neyns B, Chaskis C, Sadones J, in 't Veld P (2004) Neuropathological and molecular aspects of low-grade and high-grade gliomas. Acta Neurol Belg 104:148–153
Funding
This work was supported by grants from the National Natural Science Foundation of China (81572474), Natural Science Foundation of Beijing (7152098), and the Science and Technology Development Fund Project of Traditional Chinese Medicine of Beijing (JJ2015-14).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Dong, C., Yuan, Z., Li, Q. et al. The clinicopathological and prognostic significance of TP53 alteration in K27M mutated gliomas: an individual-participant data meta-analysis. Neurol Sci 39, 1191–1201 (2018). https://doi.org/10.1007/s10072-018-3407-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10072-018-3407-1