Abstract
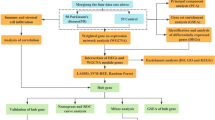
We purposed to identify underlying functional pathway cross-talk in Parkinson’s disease (PD) through Monte Carlo cross-validation analysis. Microarray data set of E-GEOD-6613 was downloaded from ArrayExpress database. First, the identification of differentially expressed genes (DEGs) was implemented, following by extracting the potential disrupted pathway enriched by DEGs. In addition, a discriminating score (DS) was computed based on the distribution of gene expression levels by quantifying their pathway cross-talk for each pair of pathways. Furthermore, random forest (RF) classification model was utilized to identify the top ten paired pathways with high AUC between PD and healthy control samples using the tenfold cross-validation method. Finally, Monte Carlo cross-validation was repeated 50 times to explore the best pairs of pathways. After quantile normalization, a total of 9331 genes with higher than 0.25-fold quantile average across all samples were obtained. Totally, 42 DEGs and 19 differential pathways enriched from DEGs were identified. We then ranked each pathway according to their AUC values, the pair of pathways, phosphatidylcholine biosynthesis I, and PPAR signaling obtained the best AUC value of 0.942. Moreover, the paired pathways of mTOR signaling and CD28 signaling in T helper cells had higher AUC value of 0.837 in five bootstraps. Two paired pathways, including phosphatidylcholine biosynthesis I and PPAR signaling, as well as mTOR signaling and CD28 signaling in T helper cells were able to accurately classify PD and healthy control samples. Significantly, these paired pathways might be underlying biomarkers for early diagnosis and therapy of PD.


Similar content being viewed by others
References
Dorsey E, Constantinescu R, Thompson J, Biglan K, Holloway R, Kieburtz K, Marshall F, Ravina B, Schifitto G, Siderowf A (2007) Projected number of people with Parkinson disease in the most populous nations, 2005 through 2030. Neurology 68:384–386
Hughes AJ, Daniel SE, Kilford L, Lees AJ (1992) Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: a clinico-pathological study of 100 cases. J Neurol Neurosur Psy 55:181–184
Lücking CB, Dürr A, Bonifati V, Vaughan J, De Michele G, Gasser T, Harhangi BS, Meco G, Denèfle P, Wood NW (2000) Association between early-onset Parkinson’s disease and mutations in the parkin gene. New Engl J Med 342:1560–1567
Murata S, Chiba T, Tanaka K (2003) CHIP: a quality-control E3 ligase collaborating with molecular chaperones. Int J Biochem Cell B 35:572–578
Edwards TL, Scott WK, Almonte C, Burt A, Powell EH, Beecham GW, Wang L, Züchner S, Konidari I, Wang G (2010) Genome-wide association study confirms SNPs in SNCA and the MAPT region as common risk factors for Parkinson disease. Ann Hum Genet 74:97–109
Spencer CC, Plagnol V, Strange A, Gardner M, Paisan-Ruiz C, Band G, Barker RA, Bellenguez C, Bhatia K, Blackburn H (2011) Dissection of the genetics of Parkinson’s disease identifies an additional association 5′ of SNCA and multiple associated haplotypes at 17q21. Hum Mol Genet 20:345–353
Tomfohr J, Lu J, Kepler TB (2005) Pathway level analysis of gene expression using singular value decomposition. BMC Bioinform 6:225
Rapaport F, Zinovyev A, Dutreix M, Barillot E, Vert JP (2007) Classification of microarray data using gene networks. BMC Bioinform 8:35
Donato M, Xu Z, Tomoiaga A, Granneman JG, MacKenzie RG, Bao R, Than NG, Westfall PH, Romero R, Draghici S (2013) Analysis and correction of crosstalk effects in pathway analysis. Genome Res 23:1885–1893
Colaprico A, Cava C, Bertoli G, Bontempi G and Castiglioni I (2015) Integrative analysis with Monte Carlo cross-validation reveals miRNAs regulating pathways cross-talk in aggressive breast cancer. BioMed Res Int 2015. doi:10.1155/2015/831314
Scherzer CR, Eklund AC, Morse LJ, Liao Z, Locascio JJ, Fefer D, Schwarzschild MA, Schlossmacher MG, Hauser MA, Vance JM (2007) Molecular markers of early Parkinson’s disease based on gene expression in blood. P Natl Acad Sci 104:955–960
Gautier L, Cope L, Bolstad BM, Irizarry RA (2004) Affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20:307–315
Robinson MD, McCarthy DJ, Smyth GK (2010) EdgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B 57:289–300
Orsetti B, Nugoli M, Cervera N, Lasorsa L, Chuchana P, Rougé C, Ursule L, Nguyen C, Bibeau F, Rodriguez C (2006) Genetic profiling of chromosome 1 in breast cancer: mapping of regions of gains and losses and identification of candidate genes on 1q. Brit J Cancer 95:1439–1447
Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA (1999) Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286:531–537
Cava C, Bertoli G, Castiglioni I (2014) Pathway-based expression profile for breast cancer diagnoses. In: Engineering in Medicine and Biology Society (EMBC), 2014 36th Annual International Conference of the IEEE. IEEE
Breiman L (2001) Random forests. Mach Learn 45:5–32
Kadowaki H, Grant MA (1995) Relationship of membrane phospholipid composition, lactosylceramide molecular species, and the specificity of CMP-N-acetylneuraminate: lactosylceramide alpha 2, 3-sialyltransferase to the molecular species composition of GM3 ganglioside. J Lipid Res 36:1274–1282
Wright MM, Howe AG, Zaremberg V (2004) Cell membranes and apoptosis: role of cardiolipin, phosphatidylcholine, and anticancer lipid. Biochem Cell Biol 26:18–26
Masuda Y, Kokubu T, Yamashita M, Ikeda H, Inoue S (1998) EGG phosphatidylcholine combined with vitamin B12 improved memory impairment following lesioning of nucleus basalis in rats. Life Sci 62:813–822
Farmer K, Smith CA, Hayley S, Smith J (2015) Major alterations of phosphatidylcholine and lysophosphotidylcholine lipids in the substantia nigra using an early stage model of Parkinson’s disease. Int J Mol Sci 16:18865–18877
Wurtman RJ, Cansev M, Sakamoto T, Ulus I (2010) Nutritional modifiers of aging brain function: use of uridine and other phosphatide precursors to increase formation of brain synapses. Nutr Rev 68:S88–S101
Marcucci H, Paoletti L, Jackowski S, Banchio C (2010) Phosphatidylcholine biosynthesis during neuronal differentiation and its role in cell fate determination. J Biol Chem 285:25382–25393
Cui Z, Houweling M (2002) Phosphatidylcholine and cell death. BBA-Mol Cell Biol Lipids 1585:87–96
Klein J (2000) Membrane breakdown in acute and chronic neurodegeneration: focus on choline-containing phospholipids. J Neural Transm 107:1027–1063
Treede I, Braun A, Sparla R, Kühnel M, Giese T, Turner JR, Anes E, Kulaksiz H, Füllekrug J, Stremmel W, Griffiths G, Ehehalt R (2007) Anti-inflammatory effects of phosphatidylcholine. J Biol Chem 282:27155–27164
Qin Z, Zhu H, Hu Y (2009) Effects of lysophosphatidylcholine on β-amyloid-induced neuronal apoptosis. Acta Pharmacol Sin 30:388–395
Wahli W, Michalik L (2012) PPARs at the crossroads of lipid signaling and inflammation. Trends Endocrin Met 23:351–363
Adibhatla RM, Hatcher JF, Dempsey RJ (2003) Phospholipase A2, hydroxyl radicals, and lipid peroxidation in transient cerebral ischemia. Antioxid Redox Sign 5:647–654
Kreisler A, Gelé P, Wiart JF, Lhermitte M, Destée A, Bordet R (2007) Lipid-lowering drugs in the MPTP mouse model of Parkinson’s disease: fenofibrate has a neuroprotective effect, whereas bezafibrate and HMG-CoA reductase inhibitors do not. Brain Res 1135:77–84
Moraes LA, Piqueras L, Bishop-Bailey D (2006) Peroxisome proliferator-activated receptors and inflammation. Pharmacol Ther 110:371–385
Ravnsjkjaer K, Frigerio F, Boegesen M, Nielsen T, Maechler P, Mandrup S (2010) PPAR-γ is a fatty acid sensor that enhances mitochondrial oxidation in insulin-secreting cells and protects against fatty acid-induced dysfunction. J Lipid Res 51:1370–1379
Taetzsch T, Block ML (2013) Pesticides, microglial NOX2, and Parkinson’s disease. J Biochem Molecular Toxic 27:137–149
Zheng B, Liao Z, Locascio JJ, Lesniak KA, Roderick SS, Watt ML, Eklund AC, Zhang-James Y, Kim PD, Hauser MA et al (2010) PGC-1α, a potential therapeutic target for early intervention in Parkinson’s disease. Sci Transl Med 2:52–73
Winklhofer KF, Haass C (2010) Mitochondrial dysfunction in Parkinson’s disease. Biochim Biophys Acta 1802:29–44
Benner EJ, Banerjee R, Reynolds AD, Sherman S, Pisarev VM, Tsiperson V, Nemachek C, Ciborowski P, Przedborski S, Mosley RL (2008) Nitrated alpha-synuclein immunity accelerates degeneration of nigral dopaminergic neurons. PLoS One 3:e1376
Miklossy J, Doudet D, Schwab C, Yu S, McGeer E, McGeer P (2006) Role of ICAM-1 in persisting inflammation in Parkinson disease and MPTP monkeys. Exp Neurol 197:275–283
Lenschow DJ, Walunas TL, Bluestone JA (1996) CD28/B7 system of T cell costimulation. Ann Rev Immunol 14:233–258
Yi-qun Z, Lorre K, de Boer M, Ceuppens JL (1997) B7-blocking agents, alone or in combination with cyclosporine A, induce antigen-specific anergy of human memory T cells. J Immunol 158:4734–4740
Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, Xiao B, Worley PF, Powell JD (2011) The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol 12(4):295–303
Hay N, Sonenberg N (2004) Upstream and downstream of mTOR. Genes Dev 18:1926–1945
Colombetti S, Basso V, Mueller DL, Mondino A (2006) Prolonged TCR/CD28 engagement drives IL-2-independent T cell clonal expansion through signaling mediated by the mammalian target of rapamycin. J Immunol 176:2730–2738
Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N (2009) Translational control of long-lasting synaptic plasticity and memory. Neuron 61:10–26
Dyskinesia D (2009) Inhibition of mTOR signaling in Parkinson’s disease prevents L-DOPA–induced dyskinesia. Sci Sign 2:ra36
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared that no conflicts of interest exist.
Rights and permissions
About this article
Cite this article
Li, T., Tang, W. & Zhang, L. Monte Carlo cross-validation analysis screens pathway cross-talk associated with Parkinson’s disease. Neurol Sci 37, 1327–1333 (2016). https://doi.org/10.1007/s10072-016-2595-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10072-016-2595-9