Abstract
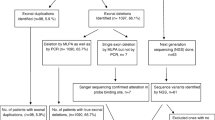
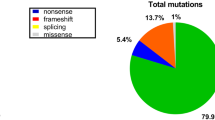
Duchenne muscular dystrophy (DMD) and Becker muscular dystrophy (BMD) are the most frequent muscular dystrophies. Present study aimed to determine the frequency of dystrophin gene alterations in Iranian DMD/BMD patients using molecular techniques. 146 Iranian DMD/BMD patients have been analyzed using two devised sets of multiplex polymerase chain reaction (M-PCR) followed by multiple ligation-dependent probe amplification (MLPA). Two isolated DMD and BMD patients were analyzed by DNA sequencing. 30.9 % of patients had single-exon deletion while group and contiguous exon deletions were identified in 41 % of the patients. The most numerous exon deletions included exons 45–50 and were identified in the first M-PCR set. Deletion detection rate was 99 % in first M-PCR set and remaining deletions (1 %) were identified in the second M-PCR set. MLPA analysis showed that there were two exons 3–5 and 41–43 duplications (1.4 %) in a BMD and a DMD patient, respectively. Two nonsense mutations including c.633dupA and c.6283 C>T were, respectively, found in a DMD and BMD patient in which c.633dupA has not ever been reported in DMD mutation database and was pathogenic mutation. Besides the report of frequency of dystrophin gene alteration in a subset of Iranian DMD/BMD patients, it was revealed that the proposed M-PCR protocol can be useful in the initial step of molecular diagnosis of DMD/BMD. Exon sequencing would be the final step in determining the mutation status of DMD/BMD patients following MLPA.

Similar content being viewed by others
Abbreviations
- ABD1:
-
Actin-binding domain 1
- BMD:
-
Becker muscular dystrophy
- CPK:
-
Creatine phosphokinase
- DMD:
-
Duchene muscular dystrophy
- EMG:
-
Electromyography
- FISH:
-
Fluorescent in situ hybridization
- IHC:
-
Immunohistochemistry
- M-PCR:
-
Multiplex polymerase chain reaction
- MAPH:
-
Multiplex amplifiable probe hybridization
- PND:
-
Prenatal diagnosis
- RPA:
-
Relative peak area
References
Landfeldt E et al (2014) The burden of Duchenne muscular dystrophy: an international, cross-sectional study. Neurology 83(6):529–536
Mampel A et al (2011) Numeric alterations in the dys gene and their association with clinical features. Medicina (B Aires) 71(2):151–157
Lee BL et al (2012) Genetic analysis of dystrophin gene for affected male and female carriers with Duchenne/Becker muscular dystrophy in Korea. J Korean Med Sci 27(3):274–280
Hermanova M et al (2002) Detection of carriers of Duchenne and Becker muscular dystrophy using the fluorescence in situ hybridization method. Cesk Patol 38(1):18–23
Velazquez-Wong AC et al (2008) Identification of duchenne muscular dystrophy female carriers by fluorescence in situ hybridization and RT-PCR. Genet Test 12(2):221–223
Chamberlain JS et al (1988) Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res 16(23):11141–11156
Stockley TL et al (2006) Strategy for comprehensive molecular testing for Duchenne and Becker muscular dystrophies. Genet Test 10(4):229–243
Nishiyama A et al (2008) Dystrophin nonsense mutations can generate alternative rescue transcripts in lymphocytes. Ann Hum Genet 72(6):717–724
Lim BC et al (2011) Genetic diagnosis of Duchenne and Becker muscular dystrophy using next-generation sequencing technology: comprehensive mutational search in a single platform. J Med Genet 48(11):731–736
Odinokova ON et al (1996) Deletion analysis of the dystrophin gene in patients with Duchenne’s muscular dystrophy in Tajikistan. Genetika 32(10):1392–1395
Mah JK et al (2011) A population-based study of dystrophin mutations in Canada. Can J Neurol Sci 38(3):465–474
Walmsley GL et al (2010) A duchenne muscular dystrophy gene hot spot mutation in dystrophin-deficient Cavalier King Charles Spaniels is amenable to exon 51 skipping. PLoS ONE 5(1):e8647
Hassan MJ et al (2008) Intragenic deletions in the dystrophin gene in 211 Pakistani Duchenne muscular dystrophy patients. Pediatr Int 50(2):162–166
Melis MA et al (1998) Elevation of serum creatine kinase as the only manifestation of an intragenic deletion of the dystrophin gene in three unrelated families. Eur J Paediatr Neurol 2(5):255–261
Muntoni F, Torelli S, Ferlini A (2003) Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol 2(12):731–740
Henderson DM, Lee A, Ervasti JM (2010) Disease-causing missense mutations in actin binding domain 1 of dystrophin induce thermodynamic instability and protein aggregation. Proc Natl Acad Sci USA 107(21):9632–9637
Prior TW et al (1993) A missense mutation in the dystrophin gene in a Duchenne muscular dystrophy patient. Nat Genet 4(4):357–360
Curtis A, Haggerty D (2001) Deletion and duplication analysis in males affected with Duchenne or Becker muscular dystrophy. Methods Mol Med 43:53–84
Khordadpoor-Deilamani F et al (2011) Dystrophin gene mutation analysis in Iranian males and females using multiplex polymerase chain reaction and multiplex ligation-dependent probe amplification methods. Genet Test Mol Biomarkers 15(12):893–899
Fenollar-Cortes M et al (2008) Two non-contiguous duplications in the DMD gene in a Spanish family. J Neurogenet 22(1):93–101
Li H et al (2009) Combining approach with multiplex PCR and MLPA to detect deletion and duplication in DMD patients, carriers, and prenatal diagnosis. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 26(3):318–322
Janssen B et al (2005) MLPA analysis for the detection of deletions, duplications and complex rearrangements in the dystrophin gene: potential and pitfalls. Neurogenetics 6(1):29–35
Beggs AH et al (1990) Detection of 98 % of DMD/BMD gene deletions by polymerase chain reaction. Hum Genet 86(1):45–48
Hwa HL et al (2007) Multiplex ligation-dependent probe amplification identification of deletions and duplications of the Duchenne muscular dystrophy gene in Taiwanese subjects. J Formos Med Assoc 106(5):339–346
Kwiatkowski M, Nilsson M, Landegren U (1996) Synthesis of full-length oligonucleotides: cleavage of apurinic molecules on a novel support. Nucleic Acids Res 24(23):4632–4638
Malayeri FA et al (2011) Detection of Duchenne/Becker muscular dystrophy carriers in a group of Iranian families by linkage analysis. Acta Med Iran 49(3):142–148
Nouri N et al (2014) Evaluation of multiplex ligation-dependent probe amplification analysis versus multiplex polymerase chain reaction assays in the detection of dystrophin gene rearrangements in an Iranian population subset. Adv Biomed Res 3:72
van Deutekom JC et al (2007) Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med 357(26):2677–2686
Acknowledgments
There is no commercial or proprietary interest in any drug, device, or equipment mentioned in this article.
Conflict of interest
No conflict of interest.
Ethical standard
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zamani, G.R., Karami, F., Mehdizadeh, M. et al. Analysis of dystrophin gene in Iranian Duchenne and Becker muscular dystrophies patients and identification of a novel mutation. Neurol Sci 36, 2011–2017 (2015). https://doi.org/10.1007/s10072-015-2290-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10072-015-2290-2