Abstract
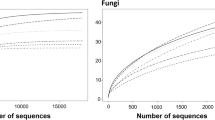
Gbodo is a traditional Nigerian fermented dried yam food using tubers or slices. Identification of microbes using conventional methods reveals only a fraction of the true microbial community. Forty-five fungal isolates associated with gbodo at 0, 6, 12, 18, and 24 h of steeping/fermentation were identified using molecular techniques. Ribosomal RNA-ITS fragments of genomic DNA were amplified using ITS1 and ITS4 primers and subjected to nuclecotide sequence determination. Meyerozyma gulliermondii represented 56–78% of isolates during 24 h of yam steeping. Sixty-seven percent of all identified isolates were members of the genera Meyerozyma and Pichia. The biodiversity index of fungal isolates increased from 0.33 at 0 h to 0.56 at 12 h. M. guilliermondii exhibited a resilient presence in isolates.
Similar content being viewed by others
References
FAO. Food Production, Food and Agricultural Organisation, Rome, Italy. p. 61 (2012)
Akissoe NH, Hounhouigan JD, Mestres C, Nago M. How blanching and drying affect the colour and functional characteristics of yam (Dioscorea cayenensis-rotundata) flour. Food Chem. 82: 257–264 (2003)
Kordylas JM. Processing and Preservation of Tropical and Subtropical Foods. Macmillan Publishers Ltd., London and Basingstoke, UK. pp. 49–71 (1990)
Akissoe NH, Hounhouigan JD, Bricas N, Vernier P, Nago MC, Olorunda OA. Physical, chemical and sensory evaluation of dried yam (Dioscorea rotundata) tubers, flour, and amala, a flour-drived product. Trop. Sci. 41: 151–155 (2001)
Babajide JM, Henshaw FO, Oyewole OB. Effect of yam variety on the pasting properties and sensory attributes of traditional dry-yam slices and their products. J. Food Quality 31: 295–305 (2008)
Onayemi O, Potter NN. Preparation and storage properties of drum dried white yam (Dioscorea rotundata Poir) flakes. J. Food Sci. 39: 559–562 (1974)
Achi OK, Akubor PI. Microbiological characterization of yam fermentation for ‘Elubo’ (yam flour) production. World J. Microb. Biot. 16: 3–7 (2000)
Babajide JM, Oyewole OB, Obadina OA. An assessment of the microbiological safety of dry yam (gbodo) processed in South West Nigeria. Afr. J. Biotechnol. 5: 157–161 (2006)
Adeyanju SA, Ikotun T. Microorganisms associated with mouldiness of dried yam ships and their prevention. Nahrung 32: 777–781 (1988)
Giraffa G, Carminati D. Molecular techniques in food fermentation: Principles and applications. pp. 1–30. In: Molecular Techniques in the Microbial Ecology of Fermented Foods. Cocolin L, Ercolini D (eds). Springer, New York, NY, USA (2008)
Mokhtari M, Etebarian HR, Razavi M, Heydari A, Mirhendi H. Identification of yeasts isolated from varieties of apples and citrus using PCR-fragment size polymorphism and sequencing of ITS1-5.8S-ITS2 region. Food Biotechnol. 26: 252–265 (2012)
Alba-Lois L, Segal-Kischinevzky C. Beer and wine makers. Nat. Educ. 3: 17 (2010)
Nout MJR, Sarkar PK. Lactic acid fermentation in tropical climates. A. Van Leeuw. 76: 395–401 (1999)
International Commission on Microbiological Specifications for Foods (ICMSF). Microorganisms in Foods. 1. Their Significance and Methods of Enumeration. 2nd ed. University of Toronto Press, Toronto, Canada. p. 158 (1978)
Barnett JA, Payne RW, Yarrow D. Yeasts: Characteristics and Identification. Cambridge University Press, Cambridge, UK. p. 86 (1983)
Rogers SO, Bendich AJ. Extraction of DNA from plant, fungal, and algal tissues. pp. 1–8. In: Plant Molecular Biology Manual. Gelvin SB, Schilperoort RA (eds). Kluwer Academic Publishers, Boston, MA, USA (1994)
Hill MO. Diversity and evenness: A unifying notation and its consequences. Ecology 54: 427–432 (1973)
American museum of natural history. Curriculum collections: How to calculate a biodiversity index. Available from: http://www.amnh.org/explore/curriculum-collections/biodiversity-counts/plant-ecology/how-to-calculate-a-biodiversity-index. Accessed Sep. 19, 2013.
Ramos CL, de Almeida EG, Freire AL, Schwan RF. Diversity of bacteria and yeast in the naturally fermented cotton seed and rice beverage produced by Brazilian Amerindians. Food Microbiol. 28: 1380–1386 (2011)
Wu X, Song R. Pichia guilliermondii strain wxm69 18S ribosomal RNA gene, partial sequence; internal transcribed spacer 1, 5.8S ribosomal RNA gene, and internal transcribed spacer 2, complete sequence; and 28S ribosomal RNA gene, partial sequence. GenBank Direct Submission. HM037942 (2010)
PengY, Chi ZM. Pichia guilliermondii strain PY-14 18S ribosomal RNA gene, partial sequence; internal transcribed spacer 1, 5.8S ribosomal RNA gene, and internal transcribed spacer 2, complete sequence; and 28S ribosomal RNA gene, partial sequence. GenBank Direct Submission. DQ674353 (2006)
Yang SP, Wu ZH, Jian JC. Distribution of marine red yeasts in shrimps and the environments of shrimp culture. Curr. Microbiol. 62: 1638–1642 (2011)
Coda R, Rizzello CG, Di Cagno R, Trani A, Cardinali G, Gobbetti M. Antifungal activity of Meyerozyma guilliermondii: Identification of active compounds synthesized during dough fermentation and their effect on long-term storage of wheat bread. Food Microbiol. 33: 243–251 (2013)
Petersson S, Schnürer J. Bio-control of mold growth in highmoisture wheat stored under airtight conditions by Pichia anomala, Pichia guilliermondii and Saccharomyces cerevisiae. Appl. Environ. Microb. 61: 1027–1032 (1995)
Rokas A. The effect of domestication on the fungal proteome. Trends Genet. 25: 60–63 (2009)
Belloch C, Querol A, Garcýa MD, Barrio E. Phylogeny of the genus Kluyveromyces inferred from the mitochondrial cytochrome-c oxidase II gene. Int. J. Syst. Evol. Micr. 50: 405–416 (2000)
Gryganskyi AP, Lee SC, Litvintseva AP, Smith ME, Bonito G, Porter TM, Anishchenko IM, Heitman J, Vilgalys R. Structure, function, and phylogeny of the mating locus in the Rhizopus oryzae complex. PLoS ONE 5: 0015273 (2010)
Nwosu VC, Oyeka CA. Microbiological succession occurring during fermentation of Ogi–an african breakfast cereal. J. Elisha Mitch. Sci. S. 114: 190–198 (1998)
Akinrele IA. Fermentation studies on maize during preparation of traditional African starch-cake food. J. Sci. Food Agr. 21: 619–625 (1970)
Babajide JM, Babajide SO, Oyewole OB. Survey of traditional dryyam slices (gbodo) processing operations in Southwest, Nigeria. Am.-Eurasian J. Sustain. Agric. 1: 45–49 (2007)
Zhang DP, Spadaro D, Valente S, Garibaldi A, Gullino ML. Cloning, characterization and expression of an exo-1,3-ß-glucanase gene from the antagonistic yeast, Pichia guilliermondii strain M8 against grey mold on apples. Biol. Control 59: 284–293 (2011)
Schirmer-Michel AC, Flores SH, Hertz PF, Matos GS, Ayub MA. Production of ethanol from soybean hull hydrolysate by osmotolerant Candida guilliermondii NRRL Y-2075. Bioresource Technol. 99: 2898–2904 (2008)
Wah TT, Walaisri S, Assavanig A, Niamsiri N, Lertsiri S. Coculturing of Pichia guilliermondii enhanced volatile flavor compound formation by Zygosaccharomyces rouxii in the model system of Thai soy sauce fermentation. Int. J. Food Microbiol. 160: 282–289 (2013)
Rodrigues RC, Sene L, Matos GS, Roberto IC, Pessoa A Jr, Felipe MG. Enhanced xylitol production by precultivation of Candida guilliermondii cells in sugarcane bagasse hemicellulosic hydrolysate. Curr. Microbiol. 53: 53–59 (2006)
Abbas CA, Sibirny AA. Genetic control of biosynthesis and transport of riboflavin and flavin nucleotides and construction of robust biotechnological producers. Microbiol. Mol. Biol. R. 75: 321–360 (2011)
Guo C, Zhao C, He P, Lu D, Shen A, Jiang N. Screening and characterization of yeasts for xylitol production. J. Appl. Microbiol. 101: 1096–1104 (2006)
Alka H, Sharma GD. Ethanol production from native yeast isolated from local cultivars of Musa sp. World J. Sci. Technol. 2: 113–117 (2012)
Savini V, Catavitello C, Onofrillo D, Masciarelli G, Astolfi D, Balbinot A, Febbo F, D’Amario C, D’Antonio D. What do we know about Candida guilliermondii? A voyage throughout past and current literature about this emerging yeast. Mycoses 54: 434–441 (2011)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Babajide, J.M., Maina, S., Kiawa, B. et al. Identification of fungal isolates from steeped yam (Gbodo): Predominance of Meyerozyma guilliermondii . Food Sci Biotechnol 24, 1041–1047 (2015). https://doi.org/10.1007/s10068-015-0133-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10068-015-0133-9