Abstract
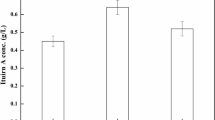
Poly-ɛ-l-lysine (ɛ-PL) biosynthesis was investigated using the resting cell culture technique and nutritional parameters were optimized with response surface methodology (RSM) and an artificial neural network (ANN). ɛ-PL production in resting cell cultures of Streptomyces noursei NRRL 5126 was compared using RSM and ANN optimization techniques. The predicted ɛ-PL yield of 924.65 mg/L using ANN simulation was in better agreement with validation experimental results of 918.35±7.56 mg/L than RSM simulation results of 966.24 mg/L. The optimized medium consisted of 3% glucose, 1% ammonium sulphate, and 5 mM citric acid in both a shake flask and a 5 L bioreactor. The shake flask ɛ-PL production as 1.0 g/L and bioreactor production as 2.36 g/ L was observed. The ANN predictive model was better than the RSM predictive model during nonlinear behavior of the system.
Similar content being viewed by others
References
Shima S, Matsuoka H, Iwamoto T, Sakai H. Antimicrobial action of ɛ-poly-l-lysine. J. Antibiot. 37: 1449–1455 (1984)
Nishikawa M, Ogawa K. Distribution of microbes producing antimicrobial ɛ-poly-l-lysine polymers in soil microflora determined by a novel method. Appl. Environ. Microbiol. 68: 3575–3581 (2002)
Hiraki J. ɛ-Polylysine, its development and utilization. Fine Chem. 29: 18–25 (2000)
Shih IL, Shen MH, Van YT. Microbial synthesis of poly(ɛ-lysine) and its various applications. Bioresour. Technol. 97: 1148–1159 (2006)
Wang Z, Zhao F, Chen D, Li D. Biotransformation of phytosterol to produce androsta-diene-dione by resting cells of mycobacterium in cloud point system. Process Biochem. 41: 557–561 (2006)
Bankar SB, Singhal RS. Metabolic precursors enhance the production of poly-ɛ-lysine by Streptomyces noursei NRRL 5126. Eng. Life Sci. 11: 253–258 (2011)
Cotter JL, Chinn MS, Grunden AM. Ethanol and acetate production by Clostridium ljungdahlii and Clostridium autoethanogenum using resting cells. Bioprocess Biosyst. Eng. 32: 369–380 (2009)
Mutafov S, Angelova B, Avramova T, Boyadjieva L, Dimova I. The inducibility of activity in resting Rhodococcus sp. cells. Process Biochem. 32: 585–589 (1997)
Zhilong W, Fengsheng Z, Daijie C, Daotang L. Biotransformation of phytosterol to produce androsta-diene-dione by resting cells of Mycobacterium in cloud point system. Process Biochem. 41: 557–561 (2006)
Bankar SB, Bule MV, Singhal RS, Anathanarayan L. Optimization of Aspergillus niger fermentation for the production of glucose oxidase. Food Bioprocess Technol. 2: 344–352 (2009)
Kagliwal LD, Survase SA, Singhal RS. A novel medium for the production of cephamycin C by Nocardia lactamdurans using solidstate fermentation. Bioresour. Technol. 100: 2600–2606 (2009)
Rao JK, Chul-Ho K, Rhee SK. Statistical optimization of medium for the production of recombinant hirudin from Saccharomyces cerevisiae using response surface methodology. Process Biochem. 35: 639–647 (2000)
Xiong YH, Liu J-Z, Song H-Y, Ji L-N. Enhanced production of extracellular ribonuclease from Aspergillus niger by optimization of culture conditions using response surface methodology. Biochem. Eng. J. 21: 27–32 (2004)
Patil SV, Jayaraman VK, Kulkarni BD. Optimization of media by evolutionary algorithms for production of polyols. Appl. Biochem. Biotechnol. 102: 119–128 (2002)
Desai KM, Survase SA, Saudagar PS, Lele SS, Singhal RS. Comparison of artificial neural network (ANN) and response surface methodology (RSM) in fermentation media optimization: Case study of fermentative production of scleroglucan. Biochem. Eng. J. 41: 266–273 (2008)
Baishan F, Hongwen C, Xiaolan X, Ning W, Zongding H. Using genetic algorithms coupling neural networks in a study of xylitol production medium optimization. Process Biochem. 38: 979–985 (2003)
Bankar SB, Singhal RS. Optimization of poly-ɛ-lysine production by Streptomyces noursei NRRL 5126. Bioresour. Technol. 101: 8370–8375 (2010)
Bankar SB, Singhal RS. Improved poly-ɛ-lysine biosynthesis using Streptomyces noursei NRRL 5126 by controlling dissolved oxygen during fermentation. J. Microbiol. Biotechnol. 21: 652–658 (2011)
Shen WC, Yang D, Ryser HJ. Calorimetric determination of microgram quantities of polylysine by trypan blue precipitation. Anal. Biochem. 142: 521–524 (1984)
Bok SH, Demain AL. An improved calorimetric assay for polyols. Anal. Biochem. 81: 18–20 (1977)
Kalil SJ, Maugeri F, Rodrigues MI. Response surface analysis and simulation as a tool for bioprocess design and optimization. Process Biochem. 35: 539–550 (2000)
Santelli RE, Bezerra MA, SantAna OD, Cassella RJ, Ferreira SLC. Multivariate technique for optimization of digestion procedure by focussed microwave system for determination of Mn, Zn and Fe in food samples using FAAS. Talanta 68: 1083 (2006)
Sivakumar M, Annadurai G, Mohan D. Studies on Box-Behnken design experiments cellulose acetate-polyurethane ultrafiltration membranes. Bioprocess Eng. 21: 65–68 (1999)
Lancashire LJ, Lemetre C, Ball GR. An introduction to artificial neural networks in bioinformatics-application to complex microarray and mass spectrometry datasets in cancer studies. Briefings Bioinf. 10: 315–329 (2009)
Nagata Y, Hoong CK. Optimization of a fermentation medium using neural networks and genetic algorithms. Biotechnol. Lett. 25: 1837–1842 (2003)
Baughman DR, Liu YA. Neural Networks in Bioprocessing and Chemical Engineering. Academic Press, San Diego, CA, USA (1995)
Leveronea MR, Owenb TC, Tieder FS, Stewartc GJ, Lim DV. Resting-cell dehydrogenase assay measuring a novel water soluble formazan detects catabolic differences among cells. J. Microbiol. Meth. 25: 49–55 (1996)
Kuwata A, Hama T, Takahashi M. Ecophysiological characterization of two life-forms, resting spores and resting cells of a marine planktonic diatom, Chaetoceros pseudo-curvisetus, formed under nutrient depletion. Mar. Ecol. Prog. Ser. 102: 245–255 (1993)
Yoshida T, Nagasawa T. ɛ-Poly-l-lysine: Microbial production, biodegradation and application potential. Appl. Microbiol. Biotechnol. 62: 21–26 (2003)
Kahar P, Iwata T, Hiraki J, Park YE, Okabe M. Enhancement of ɛ-polylysine production by Streptomyces albulus strain 410 using pH control. J. Biosci. Bioeng. 91: 190–194 (2001)
Shima S, Sakai H. Poly-l-lysine produced by Streptomyces. Part II. Taxonomy and fermentation studies. Agr. Biol. Chem. 45: 2497–2502 (1981)
James S, Legge R, Buddman H. Comparative study of black box and hybrid estimation methods in fed batch fermentation. J. Process Control. 12: 113–121 (2002)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bankar, S., Dhumal, V., Bhotmange, D. et al. Empirical predictive modelling of poly-ɛ-lysine biosynthesis in resting cells of Streptomyces noursei . Food Sci Biotechnol 23, 201–207 (2014). https://doi.org/10.1007/s10068-014-0027-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10068-014-0027-2