Abstract
Objective
Polymyositis (PM) is a chronic autoimmune connective tissue disease whose pathogenic mechanisms are unclear. This study aimed to identify the main genes and functionally enriched pathways involved in PM using weighted gene coexpression network analysis (WGCNA).
Methods
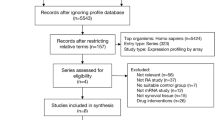
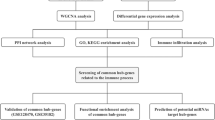
To identify the candidate genes of PM, microarray datasets GSE128470, GSE3112, GSE39454 and GSE125977 were obtained from the Gene Expression Omnibus database. The gene network of GSE128470 was constructed, and WGCNA was used to divide genes into different modules. Subsequently, Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analyses were applied to the most PM-related modules. The datasets were used to verify the expression profile and diagnostic capabilities of the hub genes. Additionally, gene set enrichment analysis (GSEA) was carried out. Moreover, gene signatures were then used as a search query to explore the connectivity map (CMap).
Results
A weighted gene coexpression network was constructed, and the genes were divided into 66 modules. The enriched functions and candidate pathway modules included interferon-γ, type I interferon, cellular response to interferon-γ, neutrophil activation, neutrophil degranulation, neutrophil-mediated immunity and neutrophil activation involved in the immune response. A total of 22 hub genes were identified. The Mann–Whitney U test was performed on these 22 genes using the three datasets of muscle samples and one dataset of whole blood samples, and two genes significantly differentially expressed in all datasets were obtained: VCAM1 and LY96. Receiver operating characteristic curve analysis determined that VCAM1 and LY96 gene expression can distinguish PM from healthy controls (the area under the curve [AUC] was greater than 0.75). Logistic regression analysis was performed on the combination of LY96 and VCAM1. The accuracy, sensitivity, specificity, and AUC of the combination reached 1.0. GSEA of VCAM1 and LY96 revealed their relation to ‘inflammatory response’, ‘TNF-α signalling via NF-κB’, ‘complement’ and ‘myogenesis’. CMap research revealed a few compounds with the potential to counteract the effects of the dysregulated molecular signature in PM.
Conclusions
We used WGCNA to observe all aspects of PM, which helped to elucidate the molecular mechanisms of PM onset and progression and provide candidate targets for the diagnosis and treatment of PM.
Key Points • Four microarray datasets were analysed in patients with polymyositis and healthy controls, and VCAM1 and LY96 were significant genes in all datasets. • GSEA of VCAM1 and LY96 revealed that they were mainly related to ‘inflammatory response’, ‘TNF-α signalling via NF-κB’, ‘complement’ and ‘myogenesis’. • CMap found a few compounds such as dimethyloxalylglycine and HNMPA-(AM)3 with the potential to counteract the effects of the dysregulated molecular signature in PM. |








Similar content being viewed by others
References
Dalakas MC, Hohlfeld R (2003) Polymyositis and dermatomyositis. Lancet 362:971–982
Dobloug C, Garen T, Bitter H, Stjärne J, Stenseth G, Grøvle L et al (2015) Prevalence and clinical characteristics of adult polymyositis and dermatomyositis; data from a large and unselected Norwegian cohort. Ann Rheum Dis 74:1551–1556
Bernatsky S, Joseph L, Pineau C, Bélisle P, Boivin J, Banerjee D et al (2009) Estimating the prevalence of polymyositis and dermatomyositis from administrative data: age, sex and regional differences. Ann Rheum Dis 68:1192–1196
Venalis P, Lundberg IE (2014) Immune mechanisms in polymyositis and dermatomyositis and potential targets for therapy. Rheumatology 53:397–405
Chen S, Wang Q, Wu Z, Li Y, Li P, Sun F et al (2014) Genetic association study of TNFAIP3, IFIH1, IRF5 polymorphisms with polymyositis/dermatomyositis in Chinese Han population. PLoS One 9:e110044
Heckmann J, Pillay K, Hearn A, Kenyon C (2010) Polymyositis in African HIV-infected subjects. Neuromuscular Disord 20:735–739
Sugiura T, Kawaguchi Y, Goto K, Hayashi Y, Tsuburaya R, Furuya T et al (2012) Positive association between STAT4 polymorphisms and polymyositis/dermatomyositis in a Japanese population. Ann Rheum Dis 71:1646–1650
Kohsaka H, Mimori T, Kanda T, Shimizu J, Sunada Y, Fujimoto M et al (2019) Treatment consensus for management of polymyositis and dermatomyositis among rheumatologists, neurologists and dermatologists. Mod Rheumatol 29:1–19
Vlekkert J, Hoogendijk J, De Haan R, Algra A, van der Tweel I, Van Der Pol W et al (2010) Oral dexamethasone pulse therapy versus daily prednisolone in sub-acute onset myositis, a randomised clinical trial. Neuromuscular Disord 20:382–389
Tjärnlund A, Tang Q, Wick C, Dastmalchi M, Mann H, Studýnková JT et al (2018) Abatacept in the treatment of adult dermatomyositis and polymyositis: a randomised, phase IIb treatment delayed-start trial. Ann Rheum Dis 77:55–62
Fasano S, Gordon P, Hajji R, Loyo E, Isenberg DA (2017) Rituximab in the treatment of inflammatory myopathies: a review. Rheumatology 56:26–36
Babaoglu H, Varan O, Atas N, Satis H, Salman R, Tufan A (2019) Tofacitinib for the treatment of refractory polymyositis. J Clin Rheumatol 25:e141–e142
Kawasumi H, Gono T, Kawaguchi Y, Kuwana M, Kaneko H, Katsumata Y et al (2016) Clinical manifestations and myositis-specific autoantibodies associated with physical dysfunction after treatment in polymyositis and dermatomyositis: an observational study of physical dysfunction with myositis in Japan. BioMed Res Int 2016:9163201
Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559
Sezin T, Vorobyev A, Sadik CD, Zillikens D, Gupta Y, Ludwig R (2018) Gene expression analysis reveals novel shared gene signatures and candidate molecular mechanisms between pemphigus and systemic lupus erythematosus in CD4+ T cells. Front Immunol 8:1992
Wickham R (2016) ggplot2: elegant graphics for data analysis. springer
Saito R, Smoot ME, Ono K, Ruscheinski J, Wang P-L, Lotia S et al (2012) A travel guide to Cytoscape plugins. Nat Methods 9:1069
Chin C-H, Chen S-H, Wu H-H, Ho C-W, Ko M-T, Lin C-Y (2014) cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 8:S11
Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C et al (2011) pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12:1–8
Yu G, Wang L-G, Han Y, He Q-Y (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287
Walter W, Sánchez-Cabo F, Ricote M (2015) GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics 31:2912–2914
Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ et al (2006) The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313:1929–1935
Musa A, Ghoraie LS, Zhang S-D, Glazko G, Yli-Harja O, Dehmer M et al (2018) A review of connectivity map and computational approaches in pharmacogenomics. Brief Bioinform 19:506–523
Shen H, Xia L, Lu J (2012) Pilot study of interleukin-27 in pathogenesis of dermatomyositis and polymyositis: associated with interstitial lung diseases. Cytokine 60:334–337
Greenberg SA (2010) Type 1 interferons and myositis. Arthritis Res Ther 12:S4
Walsh RJ, Kong SW, Yao Y, Jallal B, Kiener PA, Pinkus JL et al (2007) Type I interferon–inducible gene expression in blood is present and reflects disease activity in dermatomyositis and polymyositis. Arthritis Rheumatol 56:3784–3792
Seto N, Torres-Ruiz JJ, Carmona-Rivera C, Pinal-Fernandez I, Pak K, Purmalek MM, et al (2020) Neutrophil dysregulation is pathogenic in idiopathic inflammatory myopathies. JCI Insight 5:e134189
Koppula BR, Pipavath S, Lewis DH (2009) Epstein-Barr virus (EBV) associated lymphoepithelioma-like thymic carcinoma associated with paraneoplastic syndrome of polymyositis: a rare tumor with rare association. Clin Nucl Med 34:686–688
Nojima T, Hirakata M, Sato S, Fujii T, Suwa A, Mimori T et al (2000) A case of polymyositis associated with hepatitis B infection. Clin Exp Rheumatol 18:86–88
Wong MH, Sockalingam S, Zain A (2011) Polymyositis associated with hepatitis B: management with entacavir and prednisolone. Int J Rheum Dise 14:e38–e41
Sallum AM, Kiss MH, Silva CA, Wakamatsu A, Vianna MA, Sachetti S et al (2006) Difference in adhesion molecule expression (ICAM-1 and VCAM-1) in juvenile and adult dermatomyositis, polymyositis and inclusion body myositis. Autoimmun Rev 5:93–100
Yan W, Chen C, Chen H (2017) Estrogen downregulates miR-21 expression and induces inflammatory infiltration of macrophages in polymyositis: role of CXCL10. Mol Neurobiol 54:1631–1641
Hamann PD, Roux BT, Heward JA, Love S, McHugh NJ, Jones SW et al (2017) Transcriptional profiling identifies differential expression of long non-coding RNAs in Jo-1 associated and inclusion body myositis. Sci Rep 7:1–10
Llano-Diez M, Fury W, Okamoto H, Bai Y, Gromada J, Larsson L (2019) RNA-Sequencing reveals altered skeletal muscle contraction, E3 ligases, autophagy, apoptosis, and chaperone expression in patients with critical illness myopathy. Skelet Muscle 9:9
Jesse T, LaChance R, Iademarco M, Dean D (1998) Interferon regulatory factor-2 is a transcriptional activator in muscle where It regulates expression of vascular cell adhesion molecule-1. J Cell Biol 140:1265–1276
Malecova B, Gatto S, Etxaniz U, Passafaro M, Cortez A, Nicoletti C et al (2018) Dynamics of cellular states of fibro-adipogenic progenitors during myogenesis and muscular dystrophy. Nat Commun 9:3670
Nagai Y, Akashi S, Nagafuku M, Ogata M, Iwakura Y, Akira S et al (2002) Essential role of MD-2 in LPS responsiveness and TLR4 distribution. Nat Immunol 3:667–672
Scirocco A, Matarrese P, Petitta C, Cicenia A, Ascione B, Mannironi C et al (2010) Exposure of toll-like receptors 4 to bacterial lipopolysaccharide (LPS) impairs human colonic smooth muscle cell function. J Cell Physiol 223:442–450
Wang Y, Qian Y, Fang Q, Zhong P, Li W, Wang L et al (2017) Saturated palmitic acid induces myocardial inflammatory injuries through direct binding to TLR4 accessory protein MD2. Nat Commun 8:13997
Fang Q, Wang J, Zhang Y, Wang L, Li W, Han J et al (2018) Inhibition of myeloid differentiation factor-2 attenuates obesity-induced cardiomyopathy and fibrosis. Biochim Biophys Acta Mol Basis Dis 1864:252–262
Kim G, Cho M, Park Y, Yoo W, Kim J, Oh H et al (2010) Expression of TLR2, TLR4, and TLR9 in dermatomyositis and polymyositis. Clin Rheumatol 29:273–279
Zhang H, He F, Shi M, Wang W, Tian X, Kang J et al (2017) Toll-like receptor 4-myeloid differentiation primary response gene 88 pathway is involved in the inflammatory development of polymyositis by mediating interferon-γ and interleukin-17A in humans and experimental autoimmune myositis mouse model. Front Neurol 8:132
Chen M, Daha MR, Kallenberg CG (2010) The complement system in systemic autoimmune disease. J Autoimmun 34:J276–J286
Li L, Chen J, Chen S, Liu C, Zhang F, Li Y (2019) Serum C1q concentration positively correlates with erythrocyte sedimentation rate in polymyositis/dermatomyositis. Ann Clin Lab Sci 49:237–241
Kuru S, Inukai A, Liang Y, Doyu M, Takano A, Sobue G (2000) Tumor necrosis factor-α expression in muscles of polymyositis and dermatomyositis. Acta Neuropathol 99:585–588
Brunasso AMG, Aberer W, Massone C (2014) New onset of dermatomyositis/polymyositis during anti-TNF-α therapies: a systematic literature review. Scientific World J 2014:179180
Schiffenbauer A, Garg M, Castro C, Pokrovnichka A, Joe G, Shrader J, et al. (2018) A randomized, double-blind, placebo-controlled trial of infliximab in refractory polymyositis and dermatomyositis. Semin Arthritis Rheu. Elsevier
Fischer HJ, Sie C, Schumann E, Witte A-K, Dressel R, van den Brandt J et al (2017) The insulin receptor plays a critical role in T cell function and adaptive immunity. J Immunol 198:1910–1920
Cummins EP, Seeballuck F, Keely SJ, Mangan NE, Callanan JJ, Fallon PG et al (2008) The hydroxylase inhibitor dimethyloxalylglycine is protective in a murine model of colitis. Gastroenterology 134:156–165
Hams E, Saunders SP, Cummins EP, O’Connor A, Tambuwala MT, Gallagher WM et al (2011) The hydroxylase inhibitor DMOG attenuates endotoxic shock via alternative activation of macrophages and IL-10 production by B-1 cells. Shock 36:295
Funding
This work was supported by the National Key R&D Program of China (2020YFA0710800), Key Program of National Natural Science Foundation of China (grant no. 81930043) and Key Project supported by Medical Science and Technology Development Foundation, Nanjing Department of Health (ZKX18011).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Disclosures
None.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 2.59 mb)
Rights and permissions
About this article
Cite this article
Wang, K., Zhu, R., Li, J. et al. Coexpression network analysis coupled with connectivity map database mining reveals novel genetic biomarkers and potential therapeutic drugs for polymyositis. Clin Rheumatol 41, 1719–1730 (2022). https://doi.org/10.1007/s10067-021-06035-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10067-021-06035-5