Abstract
Background
Lupus erythematosus is an autoimmune disease that causes damage to multiple organs ranging from skin lesions to systemic manifestations. Cutaneous lupus erythematosus (CLE) is a common type of lupus erythematosus (LE), but its molecular mechanisms are currently unknown. The study aimed to explore changes in the gene expression profiles and identify key genes involved in CLE, hoping to uncover its molecular mechanism and identify new targets for CLE.
Method
We analyzed the microarray dataset (GSE109248) derived from the Gene Expression Omnibus (GEO) database, which was a transcriptome profiling of CLE cutaneous lesions. The differentially expressed genes (DEGs) were identified, and the functional annotation of DEGs was performed with Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. Protein–protein interaction (PPI) network was also constructed to identify hub genes involved in CLE.
Result
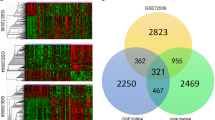
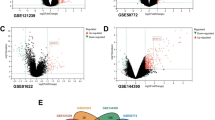
A total of 755 up-regulated DEGs and 405 down-regulated DEGs were identified. GO enrichment analysis showed that defense response to virus, immune response, and type I interferon signaling pathway were the most significant enrichment items in DEGs. The KEGG pathway analysis identified 51 significant enrichment pathways, which mainly included systemic lupus erythematosus, osteoclast differentiation, cytokine-cytokine receptor interaction, and primary immunodeficiency. Based on the PPI network, the study identified the top 10 hub genes involved in CLE, which were CXCL10, CCR7, FPR3, PPARGC1A, MMP9, IRF7, IL2RG, SOCS1, ISG15, and GSTM3. By comparison between subtypes, the results showed that ACLE had the least DEGs, while CCLE showed the most gene and functional changes.
Conclusion
The identified hub genes and functional pathways found in this study may expand our understanding on the underlying pathogenesis of CLE and provide new insights into potential biomarkers or targets for the diagnosis and treatment of CLE.
Key Points |
• The bioinformatics analysis based on CLE patients and healthy controls was performed and 1160 DEGs were identified |
• The 1160 DEGs were mainly enriched in biological processes related to immune responses, including innate immune response, type I interferon signaling pathway, interferon-γ-mediated signaling pathway, positive regulation of T cell proliferation, regulation of immune response, antigen processing, and presentation via MHC class Ib and so on |
• KEGG pathway enrichment analysis indicated that DEGs were mainly enriched in several immune-related diseases and virus infection, including systemic lupus erythematosus, primary immunodeficiency, herpes simplex infection, measles, influenza A, and so on |
• The hub genes such as CXCL10, IRF7, MMP9, CCR7, and SOCS1 may become new markers or targets for the diagnosis and treatment of CLE |







Similar content being viewed by others
Data availability
The data used for analysis in this study are available from the Gene Expression Omnibus database freely.
Code availability
The code used for analysis in this study is available from the limma packages in R language freely.
References
Chen KL, Krain RL, Werth VP (2019) Advancing understanding, diagnosis, and therapies for cutaneous lupus erythematosus within the broader context of systemic lupus erythematosus. F1000Res 8. https://doi.org/10.12688/f1000research.17787.1
Kuhn A, Wenzel J, Bijl M (2016) Lupus erythematosus revisited. Semin Immunopathol 38(1):97–112. https://doi.org/10.1007/s00281-015-0550-0
Petty AJ, Floyd L, Henderson C, Nicholas MW (2020) Cutaneous lupus erythematosus: progress and challenges. Curr Allergy Asthma Rep 20(5):12. https://doi.org/10.1007/s11882-020-00906-8
Gilliam J, Sontheimer R (1981) Distinctive cutaneous subsets in the spectrum of lupus erythematosus. J Am Acad Dermatol 4(4):471–475. https://doi.org/10.1016/s0190-9622(81)80261-7
Bertsias GK, Salmon JE, Boumpas DT (2010) Therapeutic opportunities in systemic lupus erythematosus: state of the art and prospects for the new decade. Ann Rheum Dis 69(9):1603–1611. https://doi.org/10.1136/ard.2010.135186
Fanouriakis A, Tziolos N, Bertsias G, Boumpas DT (2020) Update οn the diagnosis and management of systemic lupus erythematosus. Ann Rheum Dis. https://doi.org/10.1136/annrheumdis-2020-218272
Durcan L, O’Dwyer T, Petri M (2019) Management strategies and future directions for systemic lupus erythematosus in adults. Lancet 393(10188):2332–2343. https://doi.org/10.1016/s0140-6736(19)30237-5
Fava A, Petri M (2019) Systemic lupus erythematosus: diagnosis and clinical management. J Autoimmun 96:1–13. https://doi.org/10.1016/j.jaut.2018.11.001
Lisnevskaia L, Murphy G, Isenberg D (2014) Systemic lupus erythematosus. Lancet 384(9957):1878–1888. https://doi.org/10.1016/s0140-6736(14)60128-8
曾小峰 (2020) 2020中国系统性红斑狼疮诊疗指南. 中华内科杂志 59 (3):172–185. https://doi.org/10.3760/cma.j.issn.0578⁃1426.2020.03.002
Hejazi E, Werth V (2016) Cutaneous lupus erythematosus: an update on pathogenesis, diagnosis and treatment. Am J Clin Dermatol 17(2):135–146. https://doi.org/10.1007/s40257-016-0173-9
Grönhagen CM, Fored CM, Granath F, Nyberg F (2011) Cutaneous lupus erythematosus and the association with systemic lupus erythematosus: a population-based cohort of 1088 patients in Sweden. Br J Dermatol 164(6):1335–1341. https://doi.org/10.1111/j.1365-2133.2011.10272.x
Jiménez S, Cervera R, Font J, Ingelmo M (2003) The epidemiology of systemic lupus erythematosus. Clin Rev Allergy Immunol 25(1):3–12. https://doi.org/10.1385/criai:25:1:3
Jarukitsopa S, Hoganson DD, Crowson CS, Sokumbi O, Davis MD, Michet CJ Jr, Matteson EL, Maradit Kremers H, Chowdhary VR (2015) Epidemiology of systemic lupus erythematosus and cutaneous lupus erythematosus in a predominantly white population in the United States. Arthritis Care Res (Hoboken) 67(6):817–828. https://doi.org/10.1002/acr.22502
Petersen MP, Möller S, Bygum A, Voss A, Bliddal M (2018) Epidemiology of cutaneous lupus erythematosus and the associated risk of systemic lupus erythematosus: a nationwide cohort study in Denmark. Lupus 27(9):1424–1430. https://doi.org/10.1177/0961203318777103
Fanouriakis A, Kostopoulou M, Alunno A, Aringer M, Bajema I, Boletis JN, Cervera R, Doria A, Gordon C, Govoni M, Houssiau F, Jayne D, Kouloumas M, Kuhn A, Larsen JL, Lerstrom K, Moroni G, Mosca M, Schneider M, Smolen JS, Svenungsson E, Tesar V, Tincani A, Troldborg A, van Vollenhoven R, Wenzel J, Bertsias G, Boumpas DT (2019) 2019 update of the EULAR recommendations for the management of systemic lupus erythematosus. Ann Rheum Dis 78(6):736–745. https://doi.org/10.1136/annrheumdis-2019-215089
Osmola A, Namysł J, Jagodziński P, Prokop J (2004) Genetic background of cutaneous forms of lupus erythematosus: update on current evidence. J Appl Genet 45(1):77–86
Fischer G, Pickl W, Faé I, Anegg B, Milota S, Volc-Platzer B (1994) Association between chronic cutaneous lupus erythematosus and HLA class II alleles. Hum Immunol 41(4):280–284. https://doi.org/10.1016/0198-8859(94)90046-9
Pickering M, Fischer S, Lewis M, Walport M, Botto M, Cook H (2001) Ultraviolet-radiation-induced keratinocyte apoptosis in C1q-deficient mice. J Invest Dermatol 117(1):52–58. https://doi.org/10.1046/j.0022-202x.2001.01381.x
Werth V, Zhang W, Dortzbach K, Sullivan K (2000) Association of a promoter polymorphism of tumor necrosis factor-alpha with subacute cutaneous lupus erythematosus and distinct photoregulation of transcription. J Invest Dermatol 115(4):726–730. https://doi.org/10.1046/j.1523-1747.2000.00118.x
Järvinen T, Hellquist A, Koskenmies S, Einarsdottir E, Koskinen L, Jeskanen L, Berglind L, Panelius J, Hasan T, Ranki A, Kere J, Saarialho-Kere U (2010) Tyrosine kinase 2 and interferon regulatory factor 5 polymorphisms are associated with discoid and subacute cutaneous lupus erythematosus. Exp Dermatol 19(2):123–131. https://doi.org/10.1111/j.1600-0625.2009.00982.x
Achtman J, Werth V (2015) Pathophysiology of cutaneous lupus erythematosus. Arthritis Res Ther 17:182. https://doi.org/10.1186/s13075-015-0706-2
Wenzel J (2019) Cutaneous lupus erythematosus: new insights into pathogenesis and therapeutic strategies. Nat Rev Rheumatol 15(9):519–532. https://doi.org/10.1038/s41584-019-0272-0
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7):e47–e47. https://doi.org/10.1093/nar/gkv007
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. Gene Ontol Consort Nat Genet 25(1):25–29. https://doi.org/10.1038/75556
Ogata H, Goto S, Sato K (1999) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 27(1):29–34
Huang DW, Sherman BT, Lempicki RA (2008) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4(1):44–57. https://doi.org/10.1038/nprot.2008.211
Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16(5):284–287. https://doi.org/10.1089/omi.2011.0118
Chen HB, Boutros. PC (2011) VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12(35). https://doi.org/10.1186/1471-2105-12-35
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C (2015) STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43(D1):D447–D452. https://doi.org/10.1093/nar/gku1003
Xia J, Gill EE, Hancock RE (2015) NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat Protoc 10(6):823–844. https://doi.org/10.1038/nprot.2015.052
Liu J, Hua P, Hui L, Zhang LL, Hu Z, Zhu YW (2016) Identification of hub genes and pathways associated with hepatocellular carcinoma based on network strategy. Exp Ther Med 12(4):2109–2119. https://doi.org/10.3892/etm.2016.3599
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY (2014) cytoHubba: identifying hub objects and subnetworks from complex interactome. BMC Syst Biol 8.https://doi.org/10.1186/1752-0509-8-S4-S11
Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J (2009) ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25(8):1091–1093. https://doi.org/10.1093/bioinformatics/btp101
Shannon P (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504. https://doi.org/10.1101/gr.1239303
Subramanian A, Tamayo P, Mootha VK (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102(43):15545–15550
Robinson ES, Werth VP (2015) The role of cytokines in the pathogenesis of cutaneous lupus erythematosus. Cytokine 73(2):326–334. https://doi.org/10.1016/j.cyto.2015.01.031
Sarkar MK, Hile GA, Tsoi LC, Xing X, Liu J, Liang Y, Berthier CC, Swindell WR, Patrick MT, Shao S, Tsou P-S, Uppala R, Beamer MA, Srivastava A, Bielas SL, Harms PW, Getsios S, Elder JT, Voorhees JJ, Gudjonsson JE, Kahlenberg JM (2018) Photosensitivity and type I IFN responses in cutaneous lupus are driven by epidermal-derived interferon kappa. Ann Rheum Dis 77(11):1653–1664. https://doi.org/10.1136/annrheumdis-2018-213197
Yu C, Chang C, Zhang J (2013) Immunologic and genetic considerations of cutaneous lupus erythematosus: a comprehensive review. J Autoimmun 41:34–45. https://doi.org/10.1016/j.jaut.2013.01.007
Zucchi D, Elefante E, Calabresi E, Signorini V, Bortoluzzi A, Tani C (2019) One year in review 2019: systemic lupus erythematosus. Clin Exp Rheumatol 37:715–722
Mande P, Zirak B, Ko W-C, Taravati K, Bride KL, Brodeur TY, Deng A, Dresser K, Jiang Z, Ettinger R, Fitzgerald KA, Rosenblum MD, Harris JE, Marshak-Rothstein A (2018) Fas ligand promotes an inducible TLR-dependent model of cutaneous lupus–like inflammation. J Clin Investig 128(7):2966–2978. https://doi.org/10.1172/jci98219
Angiolillo A, Sgadari C, Taub D, Liao F, Farber J, Maheshwari S, Kleinman H, Reaman G, Tosato G (1995) Human interferon-inducible protein 10 is a potent inhibitor of angiogenesis in vivo. J Exp Med 182(1):155–162. https://doi.org/10.1084/jem.182.1.155
Sidahmed A, León A, Bosinger S, Banner D, Danesh A, Cameron M, Kelvin D (2012) CXCL10 contributes to p38-mediated apoptosis in primary T lymphocytes in vitro. Cytokine 59(2):433–441. https://doi.org/10.1016/j.cyto.2012.05.002
Romagnani P, Annunziato F, Lazzeri E, Cosmi L, Beltrame C, Lasagni L, Galli G, Francalanci M, Manetti R, Marra F, Vanini V, Maggi E, Romagnani S (2001) Interferon-inducible protein 10, monokine induced by interferon gamma, and interferon-inducible T-cell alpha chemoattractant are produced by thymic epithelial cells and attract T-cell receptor (TCR) alphabeta+ CD8+ single-positive T cells, TCRgammadelta+ T cells, and natural killer-type cells in human thymus. Blood 97(3):601–607. https://doi.org/10.1182/blood.v97.3.601
Gambichler T, Genc Z, Skrygan M, Scola N, Tigges C, Terras S, Bechara FG, Kreuter A (2012) Cytokine and chemokine ligand expression in cutaneous lupus erythematosus. Eur J Dermatol 22(3):319–323. https://doi.org/10.1684/ejd.2012.1725
Meller S, Winterberg F, Gilliet M, Müller A, Lauceviciute I, Rieker J, Neumann N, Kubitza R, Gombert M, Bünemann E, Wiesner U, Franken-Kunkel P, Kanzler H, Dieu-Nosjean M, Amara A, Ruzicka T, Lehmann P, Zlotnik A, Homey B (2005) Ultraviolet radiation-induced injury, chemokines, and leukocyte recruitment: an amplification cycle triggering cutaneous lupus erythematosus. Arthritis Rheum 52(5):1504–1516. https://doi.org/10.1002/art.21034
Iijima W, Ohtani H, Nakayama T, Sugawara Y, Sato E, Nagura H, Yoshie O, Sasano T (2003) Infiltrating CD8+ T cells in oral lichen planus predominantly express CCR5 and CXCR3 and carry respective chemokine ligands RANTES/CCL5 and IP-10/CXCL10 in their cytolytic granules: a potential self-recruiting mechanism. Am J Pathol 163(1):261–268. https://doi.org/10.1016/s0002-9440(10)63649-8
Wenzel J, Wörenkämper E, Freutel S, Henze S, Haller O, Bieber T, Tüting T (2005) Enhanced type I interferon signalling promotes Th1-biased inflammation in cutaneous lupus erythematosus. J Pathol 205(4):435–442. https://doi.org/10.1002/path.1721
Niewold T, Swedler W (2005) Systemic lupus erythematosus arising during interferon-alpha therapy for cryoglobulinemic vasculitis associated with hepatitis C. Clin Rheumatol 24(2):178–181. https://doi.org/10.1007/s10067-004-1024-2
Saban D (2014) The chemokine receptor CCR7 expressed by dendritic cells: a key player in corneal and ocular surface inflammation. Ocul Surf 12(2):87–99. https://doi.org/10.1016/j.jtos.2013.10.007
Müller G, Höpken U, Lipp M (2003) The impact of CCR7 and CXCR5 on lymphoid organ development and systemic immunity. Immunol Rev 195:117–135. https://doi.org/10.1034/j.1600-065x.2003.00073.x
López-Cotarelo P, Gómez-Moreira C, Criado-García O, Sánchez L, Rodríguez-Fernández J (2017) Beyond chemoattraction: multifunctionality of chemokine receptors in leukocytes. Trends Immunol 38(12):927–941. https://doi.org/10.1016/j.it.2017.08.004
Wenzel J, Zahn S, Mikus S, Wiechert A, Bieber T, Tüting T (2007) The expression pattern of interferon-inducible proteins reflects the characteristic histological distribution of infiltrating immune cells in different cutaneous lupus erythematosus subsets. Br J Dermatol 157(4):752–757. https://doi.org/10.1111/j.1365-2133.2007.08137.x
Wenzel J, Tüting T (2008) An IFN-associated cytotoxic cellular immune response against viral, self-, or tumor antigens is a common pathogenetic feature in “interface dermatitis.” J Invest Dermatol 128(10):2392–2402. https://doi.org/10.1038/jid.2008.96
Lauffer F, Jargosch M, Krause L, Garzorz-Stark N, Franz R, Roenneberg S, Böhner A, Mueller N, Theis F, Schmidt-Weber C, Biedermann T, Eyerich S, Eyerich K (2018) Type I immune response induces keratinocyte necroptosis and is associated with interface dermatitis. J Invest Dermatol 138(8):1785–1794. https://doi.org/10.1016/j.jid.2018.02.034
Vandooren J, Van den Steen P, Opdenakker G (2013) Biochemistry and molecular biology of gelatinase B or matrix metalloproteinase-9 (MMP-9): the next decade. Crit Rev Biochem Mol Biol 48(3):222–272. https://doi.org/10.3109/10409238.2013.770819
Gunduz K, Demireli P, Inanir I, Nese N (2006) Expression of matrix metalloproteinases (MMP-2, MMP-3, and MMP-9) and fibronectin in lichen planus. J Cutan Pathol 33(8):545–550. https://doi.org/10.1111/j.1600-0560.2006.00456.x
Ertugrul G, Keles D, Oktay G, Aktan S (2018) Matrix metalloproteinase-2 and -9 activity levels increase in cutaneous lupus erythematosus lesions and correlate with disease severity. Arch Dermatol Res 310(2):173–179. https://doi.org/10.1007/s00403-018-1811-2
Van Nguyen H, Di Girolamo N, Jackson N, Hampartzoumian T, Bullpitt P, Tedla N, Wakefield D (2011) Ultraviolet radiation-induced cytokines promote mast cell accumulation and matrix metalloproteinase production: potential role in cutaneous lupus erythematosus. Scand J Rheumatol 40(3):197–204. https://doi.org/10.3109/03009742.2010.528020
Kamizono S, Hanada T, Yasukawa H, Minoguchi S, Kato R, Minoguchi M, Hattori K, Hatakeyama S, Yada M, Morita S, Kitamura T, Kato H, Ki N, Yoshimura A (2001) The SOCS box of SOCS-1 accelerates ubiquitin-dependent proteolysis of TEL-JAK2. J Biol Chem 276(16):12530–12538. https://doi.org/10.1074/jbc.M010074200
Frantsve J, Schwaller J, Sternberg D, Kutok J, Gilliland D (2001) Socs-1 inhibits TEL-JAK2-mediated transformation of hematopoietic cells through inhibition of JAK2 kinase activity and induction of proteasome-mediated degradation. Mol Cell Biol 21(10):3547–3557. https://doi.org/10.1128/mcb.21.10.3547-3557.2001
Dong Y, Zhang Y, Xia L, Wang P, Chen J, Xu M, Liu X, Xia Y (2017) The deposition of anti-DNA IgG contributes to the development of cutaneous lupus erythematosus. Immunol Lett 191:1–9. https://doi.org/10.1016/j.imlet.2017.09.003
Swaim C, Scott A, Canadeo L, Huibregtse J (2017) Extracellular ISG15 signals cytokine secretion through the LFA-1 integrin receptor. Mol Cell 68(3):581-590.e585. https://doi.org/10.1016/j.molcel.2017.10.003
Freitas B, Scholte F, Bergeron É, Pegan S (2020) How ISG15 combats viral infection. Virus Res 286:198036. https://doi.org/10.1016/j.virusres.2020.198036
Annunziata M, Parisi M, Esposito G, Fabbrocini G, Ammendola R, Cattaneo F (2020) Phosphorylation sites in protein kinases and phosphatases regulated by formyl peptide receptor 2 signaling. Int J Mol Sci 21(11). https://doi.org/10.3390/ijms21113818
Chen K, Bao Z, Gong W, Tang P, Yoshimura T, Wang J (2017) Regulation of inflammation by members of the formyl-peptide receptor family. J Autoimmun 85:64–77. https://doi.org/10.1016/j.jaut.2017.06.012
Charos A, Reed B, Raha D, Szekely A, Weissman S, Snyder M (2012) A highly integrated and complex PPARGC1A transcription factor binding network in HepG2 cells. Genome Res 22(9):1668–1679. https://doi.org/10.1101/gr.127761.111
Ratthé C, Girard D (2004) Interleukin-15 enhances human neutrophil phagocytosis by a Syk-dependent mechanism: importance of the IL-15Ralpha chain. J Leukoc Biol 76(1):162–168. https://doi.org/10.1189/jlb.0605298
Wang S, Yang J, You L, Dai M, Zhao Y (2020) GSTM3 function and polymorphism in cancer: emerging but promising. Cancer Manag Res 12:10377–10388. https://doi.org/10.2147/cmar.S272467
Acknowledgements
We gratefully thank many researchers for providing technical assistance.
Funding
This work was supported by grants from The Affiliated Hospital of Hubei Minzu University, Enshi, Hubei Province, China.
Author information
Authors and Affiliations
Contributions
Not applicable.
Corresponding author
Ethics declarations
Ethics approval
The study was approved by the Ethics Committee of The Affiliated Hospital of Hubei Minzu University.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Disclosures
None.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gao, Zy., Su, Lc., Wu, Qc. et al. Bioinformatics analyses of gene expression profile identify key genes and functional pathways involved in cutaneous lupus erythematosus. Clin Rheumatol 41, 437–452 (2022). https://doi.org/10.1007/s10067-021-05913-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10067-021-05913-2