Abstract
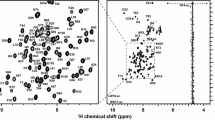
Mammalian SWI/SNF complexes are evolutionary conserved, ATP-dependent chromatin remodeling units. BAF155 in the SWI/SNF complex contains several highly conserved domains, including SANT, SWIRM, and leucine zipper domains. The biological roles of the SWIRM domain remain unclear; however, both structural and biochemical analyses of this domain have suggested that it could mediate protein-protein or protein-DNA interactions during the chromatin remodeling process. The human BAF155 SWIRM domain was cloned into the Escherichia coli expression vector pMAL-c2X and purified using affinity chromatography for structural analysis. We report the backbone 1H, 15N, and 13C resonance assignments and secondary structure of this domain using nuclear magnetic resonance (NMR) spectroscopy and the TALOS+ program. The secondary structure consists of five α-helices that form a typical histone fold for DNA interactions. Our data suggest that the BAF155 SWIRM domain interacts with nucleosome DNA (K d = 0.47 μM).
Similar content being viewed by others
References
Aasland, R., Stewart, A.F., and Gibson, T. (1996). The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem. Sci. 21, 87–88.
Barik, A., Mishra, B., Shen, L., Mohan, H., Kadam, R.M., Dutta, S., Zhang, H.Y., and Priyadarsini, K.I. (2005). Evaluation of a new copper(II)-curcumin complex as superoxide dismutase mimic and its free radical reactions. Free Radic. Biol. Med. 39, 811–822.
Barlev, N.A., Emelyanov, A.V., Castagnino, P., Zegerman, P., Bannister, A.J., Sepulveda, M.A., Robert, F., Tora, L., Kouzarides, T., Birshtein, B.K., et al. (2003). A novel human Ada2 homologue functions with Gcn5 or Brg1 to coactivate transcription. Mol. Cell. Biol. 23, 6944–6957.
Choi, Y.I., Jeon, S.H., Jang, J., Han, S., Kim, J.K., Chung, H., Lee, H.W., Chung, H.Y., Park, S.D., and Seong, R.H. (2001). Notch1 confers a resistance to glucocorticoid-induced apoptosis on developing thymocytes by down-regulating SRG3 expression. Proc. Natl. Acad. Sci. USA 98, 10267–10272.
Clapier, C.R., and Cairns, B.R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304.
Crosby, M.A., Miller, C., Alon, T., Watson, K.L., Verrijzer, C.P., Goldman-Levi, R., and Zak, N.B. (1999). The trithorax group gene moira encodes a brahma-associated putative chromatin-remodeling factor in Drosophila melanogaster. Mol. Cell. Biol. 19, 1159–1170.
Da, G., Lenkart, J., Zhao, K., Shiekhattar, R., Cairns, B.R., and Marmorstein, R. (2006). Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes. Proc. Natl. Acad. Sci. USA 103, 2057–2062.
Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. (1995). NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 6, 277–293.
Forneris, F., Binda, C., Vanoni, M., Mattevi, A., and Battaglioli, E. (2005). Histone demethylation catalysed by LSD1 is a flavin-dependent oxidative process. FEBS Lett. 579, 2203–2207.
Gui, C.Y., and Dean, A. (2003). A major role for the TATA box in recruitment of chromatin modifying complexes to a globin gene promoter. Proc. Natl. Acad. Sci. USA 100, 7009–7014.
Jeon, S.H., Kang, M.G., Kim, Y.H., Jin, Y.H., Lee, C.J., Chung, H.Y., Kwon, H., Park, S.D., and Seong, R.H. (1997). A new mouse gene, SRG3, related to the SWI3 of Saccharomyces cerevisiae, is required for apoptosis induced by glucocorticoids in a thymoma cell line. J. Exp. Med. 185, 1827–1836.
Jeong, K.W., Ko, H., Lee, S.A., Hong, E., Ko, S., Cho, H.S., Lee, W., and Kim, Y. (2013). Backbone dynamics of an atypical orphan response regulator protein, Helicobacter pylori 1043. Mol. Cells 35, 158–165.
Jung, I., Sohn, D.H., Choi, J., Kim, J.M., Jeon, S., Seol, J.H., and Seong, R.H. (2012). SRG3/mBAF155 stabilizes the SWI/SNFlike BAF complex by blocking CHFR mediated ubiquitination and degradation of its major components. Biochem. Biophys. Res. Commun. 418, 512–517.
Kim, B., Kang, S., and Kim, S.J. (2012). Differential promoter methylation and histone modification contribute to the brain specific expression of the mouse Mbu-1 gene. Mol. Cells 34, 433–437.
Ko, M. (2004). T cell receptor signaling inhibits glucocorticoidinduced apoptosis by repressing the SRG3 expression via Ras activation. J. Biol. Chem. 279, 21903–21915.
Ko, S., Kang, G.B., Song, S.M., Lee, J.G., Shin, D.Y., Yun, J.H., Sheng, Y., Cheong, C., Jeon, Y.H., Jung, Y.K., et al. (2010). Structural basis of E2-25K/UBB+1 interaction leading to proteasome inhibition and neurotoxicity. J. Biol. Chem. 285, 36070–36080.
Lee, W., Westler, W.M., Bahrami, A., Eghbalnia, H.R., and Markley, J.L. (2009). PINE-SPARKY: graphical interface for evaluating automated probabilistic peak assignments in protein NMR spectroscopy. Bioinformatics 25, 2085–2087.
Lee, C.J., Yun, J.H., Lim, S.K., and Lee, W. (2010). Solution structures and molecular interactions of selective melanocortin receptor antagonists. Mol. Cells 30, 551–556.
Nissler, L., Gebhardt, R., and Berger, S. (2004). Flavonoid binding to a multi-drug-resistance transporter protein: an STD-NMR study. Anal. Bioanal. Chem. 379, 1045–1049.
Phelan, M.L., Sif, S., Narlikar, G.J., and Kingston, R.E. (1999). Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3, 247–253.
Qian, C., Zhang, Q., Li, S., Zeng, L., Walsh, M.J., and Zhou, M.M. (2005). Structure and chromosomal DNA binding of the SWIRM domain. Nat. Struct. Mol. Biol. 12, 1078–1085.
Reisman, D., Glaros, S., and Thompson, E.A. (2009). The SWI/SNF complex and cancer. Oncogene 28, 1653–1668.
Roberts, C.W., and Orkin, S.H. (2004). The SWI/SNF complex-chromatin and cancer. Nat. Rev. Cancer 4, 133–142.
Shen, Y., Delaglio, F., Cornilescu, G., and Bax, A. (2009). TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 44, 213–223.
Sohn, D.H., Lee, K.Y., Lee, C., Oh, J., Chung, H., Jeon, S.H., and Seong, R.H. (2007). SRG3 interacts directly with the major components of the SWI/SNF chromatin remodeling complex and protects them from proteasomal degradation. J. Biol. Chem. 282, 10614–10624.
Wu, J.I., Lessard, J., and Crabtree, G.R. (2009). Understanding the words of chromatin regulation. Cell 136, 200–206.
Wuthrich, K. (1990). Protein structure determination in solution by NMR spectroscopy. J. Biol. Chem. 265, 22059–22062.
Yoneyama, M., Tochio, N., Umehara, T., Koshiba, S., Inoue, M., Yabuki, T., Aoki, M., Seki, E., Matsuda, T., Watanabe, S., et al. (2007). Structural and functional differences of SWIRM domain subtypes. J. Mol. Biol. 369, 222–238.
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Moon, S., Shin, J., Lee, D. et al. 1H, 15N, and 13C resonance assignments and secondary structure of the SWIRM domain of human BAF155, a chromatin remodeling complex component. Mol Cells 36, 333–339 (2013). https://doi.org/10.1007/s10059-013-0119-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10059-013-0119-5