Abstract:
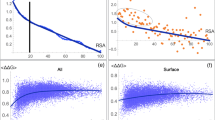
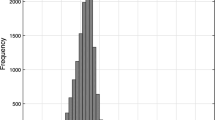
We simulate the evolution of a protein-like sequence subject to point mutations, imposing conservation of the ground state, thermodynamic stability and fast folding. Our model is aimed at describing neutral evolution of natural proteins. We use a cubic lattice model of the protein structure and test the neutrality conditions by extensive Monte Carlo simulations. We observe that sequence space is traversed by neutral networks, i.e. sets of sequences with the same fold connected by point mutations. Typical pairs of sequences on a neutral network are nearly as different as randomly chosen sequences. The fraction of neutral neighbors has strong sequence to sequence variations, which influence the rate of neutral evolution. In this paper we study the thermodynamic stability of different protein sequences. We relate the high variability of the fraction of neutral mutations to the complex energy landscape within a neutral network, arguing that valleys in this landscape are associated to high values of the neutral mutation rate. We find that when a point mutation produces a sequence with a new ground state, this is likely to have a low stability. Thus we tentatively conjecture that neutral networks of different structures are typically well separated in sequence space. This result indicates that changing significantly a protein structure through a biologically acceptable chain of point mutations is a rare, although possible, event.
Similar content being viewed by others
Author information
Authors and Affiliations
Additional information
Received 8 July 1999
Rights and permissions
About this article
Cite this article
Bastolla, U., Vendruscolo, M. & Roman, H. Structurally constrained protein evolution: results from a lattice simulation. Eur. Phys. J. B 15, 385–397 (2000). https://doi.org/10.1007/s100510051140
Issue Date:
DOI: https://doi.org/10.1007/s100510051140