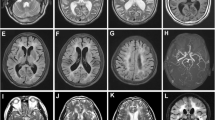
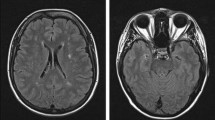
Abstract
Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) is the most common monogenic disease leading to stroke and vascular dementia. CADASIL is an inherited small blood vessel disease caused by mutations in the gene encoding the neurogenic locus notch homolog protein 3 (NOTCH3). NOTCH3 is large type I membrane receptor mainly expressed in vascular smooth muscle cells and pericytes. Most identified mutations result in insert or deletion of a cysteine residue within the EGF-like repeats. To date, some cases with a cysteine-sparing mutant have been described. Genetic analysis revealed a novel mutation in NOTCH3 in a CADASIL family. Molecular analysis revealed its potential pathogenic mechanism in causing CADASIL. In this paper, we present a Chinese family with a novel cysteine-sparing mutation in exon 3 (c.218G>C, p.G73A) of the NOTCH3 gene. Family carriers of the same mutation presented with symptoms and imaging abnormalities characteristic of CADASIL. The location of glycine 73 in between C5-C6 disulfide bond of EGF-like domain 1 shows high conservation from humans to zebra fish. It has previously been suggested that the aggregate-prone property of mutant NOTCH3 contributes to a cytotoxic effect in the pathogenic mechanism underlying CADASIL. Here, we investigated the pathogenic mechanism of the new mutation in vitro using HEK293 cells transfected with either a wild-type (WT) or c.218G>C (p.G73A) NOTCH3ECD plasmids, and we found p.G73A NOTCH3ECD was more prone to form aggregation and resistant to degradation. Moreover, the p.G73A NOTCH3ECD compromised cell viability by promoting apoptosis. Two known CADASIL mutants R133C and R75P showed similar results with G73A mutants. Our study here identified G73A as a new mutation in NOTCH3 to cause CADASIL and revealed that the G73A mutation and two known mutants R75P and R133C decreased NOTCH3 protein turnover and induced cell death.





Similar content being viewed by others
Abbreviations
- ANOVA :
-
analysis of variance
- CADASIL :
-
cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy
- CCK8 :
-
Cell Counting Kit-8
- CHD :
-
coronary heart disease
- CHX :
-
chlorhexidine
- DAPI :
-
4′,6′-diamino-2-phenylindole
- DM :
-
diabetes mellitus
- DSL :
-
Delta/Serrate/Lag-2
- ECD :
-
extracellular domain
- ECM :
-
extracellular matrix
- EGF-like repeats :
-
epidermal growth factor-like repeats
- ER :
-
endoplasmic reticulum
- FCM :
-
flow cytometry
- FITC :
-
fluorescein isothiocyanate
- FLAIR:
-
fluid-attenuated inversion recovery magnetic resonance imaging
- GOM :
-
granular osmiophilic material
- HAMA:
-
Hamilton Anxiety Scale
- HEK293 :
-
human embryonic kidney 293 cells
- ICD :
-
intracellular domain
- IGT :
-
impaired glucose tolerance
- MRI :
-
magnetic resonance imaging
- MMSE :
-
mini-mental state examination
- NOTCH3 :
-
neurogenic locus notch homolog protein 3
- PBS :
-
phosphate-buffered saline
- PI :
-
propidium iodide
- SWI :
-
susceptibility weighted imaging
- UPR :
-
unfolded protein response
- VSMCs :
-
vascular smooth muscle cells
- MH :
-
T2-weighted white matter hyperintensity
- WT :
-
wild type
References
Pantoni L (2010) Cerebral small vessel disease: from pathogenesis and clinical characteristics to therapeutic challenges. Lancet Neurol. 9(7):689–701
Chabriat H, Joutel A, Dichgans M, Tournier-Lasserve E, Bousser MG (2009) Cadasil. Lancet Neurol. 8(7):643–653
Takahashi K, Adachi K, Yoshizaki K, Kunimoto S, Kalaria RN, Watanabe A (2010) Mutations in NOTCH3 cause the formation and retention of aggregates in the endoplasmic reticulum, leading to impaired cell proliferation. Human molecular genetics. 19(1):79–89
Domenga V, Fardoux P, Lacombe P, Monet M, Maciazek J, Krebs LT, Klonjkowski B, Berrou E, Mericskay M, Li Z, Tournier-Lasserve E, Gridley T, Joutel A (2004) Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev. 18(22):2730–2735
Cognat E, Baron-Menguy C, Domenga-Denier V, Cleophax S, Fouillade C, Monet-Lepretre M et al (2014) Archetypal Arg169Cys mutation in NOTCH3 does not drive the pathogenesis in cerebral autosomal dominant arteriopathy with subcortical infarcts and leucoencephalopathy via a loss-of-function mechanism. Stroke. 45(3):842–849
Ge W, Kuang H, Wei B, Bo L, Xu Z, Xu X, Geng D, Sun M (2014) A novel cysteine-sparing NOTCH3 mutation in a Chinese family with CADASIL. PLoS One. 9(8):e104533
Louvi A, Artavanis-Tsakonas S (2012) Notch and disease: a growing field. Semin Cell Dev Biol. 23(4):473–480
Fortini ME, Bilder D (2009) Endocytic regulation of Notch signaling. Curr Opin Genet Dev. 19(4):323–328
Kopan R (2012) Notch signaling. Cold Spring Harb Perspect Biol 4(10)
Meng H, Zhang X, Yu G, Lee SJ, Chen YE, Prudovsky I, Wang MM (2012) Biochemical characterization and cellular effects of CADASIL mutants of NOTCH3. PLoS One. 7(9):e44964
Joutel A, Vahedi K, Corpechot C, Troesch A, Chabriat H, Vayssiere C et al (1997) Strong clustering and stereotyped nature of Notch3 mutations in CADASIL patients. Lancet. 350(9090):1511–1515
Monet M, Domenga V, Lemaire B, Souilhol C, Langa F, Babinet C, Gridley T, Tournier-Lasserve E, Cohen-Tannoudji M, Joutel A (2007) The archetypal R90C CADASIL-NOTCH3 mutation retains NOTCH3 function in vivo. Human molecular genetics. 16(8):982–992
Kitamoto T, Takahashi K, Takimoto H, Tomizuka K, Hayasaka M, Tabira T, Hanaoka K (2005) Functional redundancy of the Notch gene family during mouse embryogenesis: analysis of Notch gene expression in Notch3-deficient mice. Biochem Biophys Res Commun. 331(4):1154–1162
Donahue CP, Kosik KS (2004) Distribution pattern of Notch3 mutations suggests a gain-of-function mechanism for CADASIL. Genomics. 83(1):59–65
Monet-Lepretre M, Haddad I, Baron-Menguy C, Fouillot-Panchal M, Riani M, Domenga-Denier V et al (2013) Abnormal recruitment of extracellular matrix proteins by excess Notch3 ECD: a new pathomechanism in CADASIL. Brain : a journal of neurology. 136(Pt 6):1830–1845
Rutten JW, Boon EM, Liem MK, Dauwerse JG, Pont MJ, Vollebregt E, Maat-Kievit AJ, Ginjaar HB, Lakeman P, van Duinen S, Terwindt GM, Lesnik Oberstein SA (2013) Hypomorphic NOTCH3 alleles do not cause CADASIL in humans. Hum Mutat. 34(11):1486–1489
Joutel A (2011) Pathogenesis of CADASIL: transgenic and knock-out mice to probe function and dysfunction of the mutated gene, Notch3, in the cerebrovasculature. Bioessays. 33(1):73–80
Duering M, Karpinska A, Rosner S, Hopfner F, Zechmeister M, Peters N, Kremmer E, Haffner C, Giese A, Dichgans M, Opherk C (2011) Co-aggregate formation of CADASIL-mutant NOTCH3: a single-particle analysis. Human molecular genetics. 20(16):3256–3265
Opherk C, Duering M, Peters N, Karpinska A, Rosner S, Schneider E, Bader B, Giese A, Dichgans M (2009) CADASIL mutations enhance spontaneous multimerization of NOTCH3. Human molecular genetics. 18(15):2761–2767
Ihalainen S, Soliymani R, Iivanainen E, Mykkanen K, Sainio A, Poyhonen M et al (2007) Proteome analysis of cultivated vascular smooth muscle cells from a CADASIL patient. Mol Med. 13(5-6):305–314
Muino E, Gallego-Fabrega C, Cullell N, Carrera C, Torres N, Krupinski J et al (2017) Systematic review of cysteine-sparing NOTCH3 missense mutations in patients with clinical suspicion of CADASIL. International journal of molecular sciences 18(9)
Vlachakis D, Tsaniras SC, Ioannidou K, Baumann M, Kossida S (2014) A series of NOTCH3 mutations in CADASIL; insights from 3D molecular modelling and evolutionary analyses. J Mol Biochem. 3:97–105
Wollenweber FA, Hanecker P, Bayer-Karpinska A, Malik R, Bazner H, Moreton F et al (2015) Cysteine-sparing CADASIL mutations in NOTCH3 show proaggregatory properties in vitro. Stroke. 46(3):786–792
Zhu Y, Wang J, Wu Y, Wang G, Hu B (2015) Two novel mutations in NOTCH3 gene causes cerebral autosomal dominant arteriopathy with subcritical infarct and leucoencephalopathy in two Chinese families. Int J Clin Exp Pathol. 8(2):1321–1327
Wang Z, Yuan Y, Zhang W, Lv H, Hong D, Chen B, Liu Y, Luan X, Xie S, Wu S (2011) NOTCH3 mutations and clinical features in 33 mainland Chinese families with CADASIL. Journal of neurology, neurosurgery, and psychiatry. 82(5):534–539
Chen S, Ni W, Yin XZ, Liu HQ, Lu C, Zheng QJ, Zhao GX, Xu YF, Wu L, Zhang L, Wang N, Li HF, Wu ZY (2017) Clinical features and mutation spectrum in Chinese patients with CADASIL: a multicenter retrospective study. CNS Neurosci Ther. 23(9):707–716
Joutel A, Haddad I, Ratelade J, Nelson MT (2016) Perturbations of the cerebrovascular matrisome: a convergent mechanism in small vessel disease of the brain? J Cereb Blood Flow Metab. 36(1):143–157
Yamamoto Y, Craggs LJ, Watanabe A, Booth T, Attems J, Low RW, Oakley AE, Kalaria RN (2013) Brain microvascular accumulation and distribution of the NOTCH3 ectodomain and granular osmiophilic material in CADASIL. J Neuropathol Exp Neurol. 72(5):416–431
Joutel A, Andreux F, Gaulis S, Domenga V, Cecillon M, Battail N, Piga N, Chapon F, Godfrain C, Tournier-Lasserve E (2000) The ectodomain of the Notch3 receptor accumulates within the cerebrovasculature of CADASIL patients. The Journal of clinical investigation. 105(5):597–605
Ghezali L, Capone C, Baron-Menguy C, Ratelade J, Christensen S, Ostergaard Pedersen L et al (2018) Notch3(ECD) immunotherapy improves cerebrovascular responses in CADASIL mice. Ann Neurol.
Desmond DW, Moroney JT, Lynch T, Chan S, Chin SS, Mohr JP (1999) The natural history of CADASIL: a pooled analysis of previously published cases. Stroke. 30(6):1230–1233
Chabriat H, Tournier-Lasserve E, Vahedi K, Leys D, Joutel A, Nibbio A, Escaillas JP, Iba-Zizen MT, Bracard S, Tehindrazanarivelo A (1995) Autosomal dominant migraine with MRI white-matter abnormalities mapping to the CADASIL locus. Neurology. 45(6):1086–1091
Vyshka G, Kruja J (2013) Clinical variability of the cerebral autosomal-dominant arteriopathy with subcortical infarcts and leukoencephalopathy phenotype in two siblings of a large family showing the same mutation. Int Med Case Rep J. 6:59–63
Tan ZX, Li FF, Qu YY, Liu J, Liu GR, Zhou J, Zhu YL, Liu SL (2012) Identification of a known mutation in Notch 3 in familiar CADASIL in China. PLoS One. 7(5):e36590
Dziewulska D, Sulejczak D, Wezyk M (2017) What factors determine phenotype of cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL)? Considerations in the context of a novel pathogenic R110C mutation in the NOTCH3 gene. Folia Neuropathol. 55(4):295–300
Monet-Lepretre M, Bardot B, Lemaire B, Domenga V, Godin O, Dichgans M et al (2009) Distinct phenotypic and functional features of CADASIL mutations in the Notch3 ligand binding domain. Brain : a journal of neurology. 132(Pt 6):1601–1612
Adib-Samii P, Brice G, Martin RJ, Markus HS (2010) Clinical spectrum of CADASIL and the effect of cardiovascular risk factors on phenotype: study in 200 consecutively recruited individuals. Stroke. 41(4):630–634
Acknowledgements
We thank Dr. Weihong Song for his instructive suggestions.
Availability of data and materials
All data generated or analyzed during this study are included in this published article (and as supplement original data.pdf).
Funding
This study was supported by grants from NSFC (81771155) to Xiulian Sun and grants from Shandong Key Research Project (2018GSF118104) to Hongchun Wang.
Author information
Authors and Affiliations
Contributions
L. H. and X. S. designed the experiments and wrote the manuscript. W. L. collected the clinical data. L. H. performed the molecular studies. Y. L., C. S., P. W., and H. W. helped in preparing the materials. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical statements
Study protocols were approved by the Medical Ethical Committee of Qingdao Municipal Hospital (2016-002). All persons gave their informed consent prior to their inclusion in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Huang, L., Li, W., Li, Y. et al. A novel cysteine-sparing G73A mutation of NOTCH3 in a Chinese CADASIL family. Neurogenetics 21, 39–49 (2020). https://doi.org/10.1007/s10048-019-00592-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-019-00592-3