Abstract
Hereditary paroxysmal dyskinesias (PxD) are a heterogeneous group of movement disorders classified by frequency, duration, and triggers of the episodes. A young-adult onset canine PxD has segregated as an autosomal recessive trait in Soft-Coated Wheaten Terriers. The medical records and videos of episodes from 25 affected dogs were reviewed. The episodes of hyperkinesia and dystonia lasted from several minutes to several hours and could occur as often as >10/day. They were not associated with strenuous exercise or fasting but were sometimes triggered by excitement. The canine PxD phenotype most closely resembled paroxysmal non-kinesigenic dyskinesia (PNKD) of humans. Whole genome sequences were generated with DNA from 2 affected dogs and analyzed in comparison to 100 control canid whole genome sequences. The two whole genome sequences from dogs with PxD had a rare homozygous PIGN:c.398C > T transition, which predicted the substitution of an isoleucine for a highly conserved threonine in the encoded enzyme. All 25 PxD-affected dogs were PIGN:c.398T allele homozygotes, whereas there were no c.398T homozygotes among 1185 genotyped dogs without known histories of PxD. PIGN encodes an enzyme involved in the biosynthesis of glycosylphosphatidylinositol (GPI), which anchors a variety of proteins including CD59 to the cell surface. Flow cytometry of PIGN-knockout HEK239 cells expressing recombinant human PIGN with the c.398T variant showed reduced CD59 expression. Mutations in human PIGN have been associated with multiple congenital anomalies-hypotonia-seizures syndrome-1 (MCAHS1). Movement disorders can be a part of MCAHS1, but this is the first PxD associated with altered GPI anchor function.
Similar content being viewed by others
Introduction
The human paroxysmal dyskinesias (PxD) are a heterogeneous group of diseases characterized by episodes of abnormal involuntary movements [1]. The episodes may last <1 min or continue for many hours. Episode frequency may vary from <1/year to >100/day. The abnormal movements can be dystonia, chorea, athetosis, or ballism, either singly or in various combinations [2–4]. Many PxD episodes are initiated by a recognized trigger, and the type of trigger has served as a means of subclassifying PxD [1, 2, 5]. In paroxysmal exertional dyskinesia (PED), the episodes are triggered by sustained exercise. Sudden movements trigger the episodes in paroxysmal kinesigenic dyskinesia (PKD). In paroxysmal non-kinesigenic dyskinesia (PNKD), stress, fatigue, and hunger can trigger episodes in some patients, but the most consistent triggers are alcohol or caffeine consumption. Other factors considered in classification are age of onset, frequency and duration of the episodes, and response to therapy [1, 2, 5–7].
PxD may be secondary to a specific etiology such as encephalitis or stroke [8] or idiopathic. The latter can be sporadic or familial, usually with an autosomal dominant inheritance [5]. Mutations in three genes, SLC2A1, PRRT2, and PNKD (also referred to as MR-1), are well-recognized causes of PxD [5, 9–13]. SLC2A1 encodes glucose transport protein 1 (GLUT1), which facilitates glucose transfer across the blood-brain barrier. Most patients with heterozygous SLC2A1 mutations exhibit GLUT1-deficiency syndrome characterized by low CSF-to-blood glucose ratios together with epilepsy, developmental delay, microcephaly, ataxia, and PxD [14]. A minority of patients with heterozygous SLC2A1 mutations develop isolated PED [11, 15–17]. PRRT2 encodes proline-rich transmembrane protein 2, which interacts with SNAP25 and thereby influences the release of glutamate or other neurotransmitters [18]. Heterozygous PRRT2 mutations have been found in patients with several paroxysmal neurologic disorders including PKD [19]. Many PNKD patients carry heterozygous mutations in PNKD [5, 6, 13]. This gene encodes a protein that may suppress synaptic vesicle release by interacting with RIM1 and RIM2 [20]. Although SLC2A1, PNKD, and PRRT2 harbor the majority of the causal mutations for the genetically defined cases of PxD [5], a few patients have been reported to carry potentially causal mutations in other genes such as KCNMA1 [21] and PDHA1 [22]. Efforts to identify the mutations responsible for PxD in other families have not yet been successful [6].
Dogs can also develop PxD [23]. Published reports have provided clinical descriptions of primary canine PxD (cPxD) or cPxD-like diseases in several breeds including the Bichon Frise [24], Border Terrier [25], Cavalier King Charles Spaniel [26], Chinook [27], Doberman Pinscher [28], English Bulldog [29], Scottish Terrier [30], and Soft-Coated Wheaten Terrier (SCWT) [31]. Unlike the human primary PxD that are almost always dominant traits, the cPxD usually have a recessive mode of inheritance. The cPxD that occurs in the Cavalier King Charles Spaniel is typically precipitated by exercise [26]. It is caused by a homozygous 16-kb microdeletion that includes the first three exons of BCAN [32, 33], which encodes the extracellular-matrix protein brevican. The molecular genetic causes of the other cPxD have not yet been reported. We now present evidence that SCWT with cPxD have a deficiency in the biosynthesis of glycosylphosphatidyinositol (GPI) anchors due to a homozygous missense mutation in PIGN, the gene that encodes a GPI synthesis enzyme, GPI ethanolamine phosphate transferase-1.
Methods
Animals
Medical records and available videos of dyskinesia episodes were reviewed for 22 SCWT and 3 “Whoodles” (SCWT × Poodle crosses) with cPxD. The median age of onset of signs was 2.25 years. Males and females were affected equally. All dogs had normal neurologic exams between the episodes. Episode duration ranged from several minutes up to >4 h, and the frequency of episodes ranged from 1 every few days to >10/day. In six cases, stress, excitement, or loud noises were thought to precipitate some attacks, but a clear trigger was not apparent in most instances. No dogs had a history of ingestion of alcohol or caffeine. Typical episodes consisted of rapid flexion and extension of the hind limbs with varying degrees of truncal dystonia (Supplementary Video). Most frequently, the flexion alternated irregularly between limbs, but sometimes both hind limbs were off the ground simultaneously (Fig. 1). In severe episodes, the front limbs were also affected. No consistent improvement in episodes was reported with anti-epileptic drugs or benzodiazepines. The signs typically worsened with age, and six dogs were euthanized within 2 years after onset of signs due to the increasing severity of the episodes. MRIs were obtained from five affected dogs, and postmortem exams were performed on four dogs, but no abnormalities were identified in any of the examined brains.
Molecular genetics
DNA was collected from 25 cPxD-affected dogs. In addition, we used archived DNA samples from 665 cPxD-free SCWT, 388 Poodles, and 132 randomly selected dogs from other breeds to validate the identified candidate causal variant. Whole genome sequences were generated with DNA from two different cPxD-affected SCWT. PCR-based DNA libraries were used for the first whole genome sequence as previously described [34]. PCR-free DNA libraries and slightly modified procedures [35] were used for the second. Both data sets were deposited in the Sequence Read Archives (accession numbers SRX863100 and SRX863101). Previously described procedures were used for trimming adaptor sequences from the sequence reads, eliminating duplicate reads, error correction, and alignment to the CanFam3.1 canine reference genome sequence [34, 36]. Sequence variant information was uploaded to a custom PostgreSQL database, which also contained sequence variant information from the aligned whole genome sequences of normal canids or dogs with diseases other than cPxD. These other sequences served as controls for the current analysis. The University of Missouri DNA Core Facility generated 43 of the control dog genome sequences; the rest were provided by individuals at other institutions as indicated in the “Acknowledgements.”
PCR primers 5′-GCTATACTAATATTTCACCGTTC-3′ and 5′-AAAATATAGTAAGTAATACACAA-3′ were used to amplify a segment of genomic DNA containing candidate sequence variant PIGN:c.398C > T, identified from the whole genome sequences, so that the sequence variant could be verified by direct automated Sanger sequencing. An allelic discrimination assay [37] was used to genotype DNA samples from individual dogs at this sequence variant. PCR primers for this assay were 5′-TGTGGGATTTGATTCTCTTATTAATG-3′ and 5′-TCAATGACTCTTACCTTTGGCAAACATA-3′. The competing probe sequences were 5′-VIC-CCAGCTCCAT G TGTACC-MBG-3′ (reference C allele) and 5′-FAM-CCAGCTCCAT A TGTACC-MBG-3′ (variant T allele).
PIGN-knockout HEK293 cells were generated and transfected as previously described [38] with human wild-type or mutant (T133I, T133S, or T133V) PIGN complementary DNA (DNA) cloned into pME, a strong SRα promoter-driven expression vector, or pTK, a medium TK promoter-driven expression vector. These PIGN constructs had an HA epitope tag at the N-terminus. Transfection efficiencies were determined by the luciferase activity of cell lysates. After 3 days of incubation, restoration of the cell-surface expression of CD59 was evaluated by flow cytometry [38]. Levels of expressed wild-type and mutant HA-tagged PIGN in pME vector-transfected cells were determined by western blotting using an anti-HA antibody.
Results
Molecular genetics
Partial pedigree information was available for some of the affected dogs. Because no records indicated that the sires or dams of any of the affected dogs also exhibited cPxD, we assumed an autosomal recessive inheritance in our data analysis.
The first generated whole genome sequence from a cPxD-affected dog had a 21-fold average coverage of the reference genome and contained 6.9 million potential sequence variants (homozygous or heterozygous differences from the reference canine genome sequence). When this sequence variant information was first added to a custom PostgreSQL database, it also contained sequence variant information from eight control canine WGSs. The SCWT sequence variants were filtered to identify only those that fit three criteria: (1) they were predicted to alter the amino acid sequence of the gene product (including those that alter exon-splicing signals), (2) they were homozygous in the cPxD-affected dog’s whole genome sequence, and (3) they were absent from the control whole genome sequences. In the initial analysis, 418 sequence variants met these criteria. However, no plausible candidate sequence variants were identified from a review of the published biological functions and disease associations for the 302 genes that harbored these sequence variants.
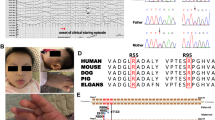
As additional control whole genome sequences were added to the database, the number of homozygous coding variants unique to the affected SCWT decreased. Nonetheless, we were still unable to recognize a potential cPxD-causing sequence variant. Thus, we generated a whole genome sequence from another cPxD-affected dog. This sequence had 18-fold average reference genome coverage and contained 5.5 million potential sequence variants. By the time that the variants from the second cPxD-affected SCWT were included in our database, it contained variant data for 100 canid sequences that could be considered to be controls for the SCWT PxD. At this time, the whole genome sequence for the first case SCWT contained 230 variants in 162 different genes that met our criteria for candidate causality (Supplementary Table 1a). The whole genome sequence for the second SCWT PxD case had 65 candidate variants in 54 different genes (Supplementary Table 1b). The higher number of variants from the first case is attributable to the many false positives among the filtered variants from the early PCR-amplified libraries. The only sequence variant common to both animals was PIGN:c.398C > T, which predicted a p. T133I substitution in PIGN, the gene product. Direct automated Sanger sequencing of PCR amplicons produced with primers spanning the PIGN:c.398C > T variant confirmed that both of the SCWT with cPxD were PIGN:c.398T homozygotes.
All 25 of our DNA samples from cPxD-affected dogs tested homozygous for the variant PIGN:c.398T allele. Twenty two of these affected homozygotes were SCWT, and the other three were Whoodles. Whoodles are marketed as the product of F1 crosses between purebred SCWT and purebred Poodles. If this was indeed the origin of the three c.398T homozygous Whoodles, it would require that the PIGN:c.398T allele had been segregating in both breeds. Thus, we genotyped individual representatives from each breed at PIGN:c.398C > T. Fifteen of the 682 control SCWT were heterozygotes; the remaining 667 were homozygous for reference allele c.398C. All 388 of the genotyped Poodles were PIGN:c.398C homozygotes, as were all 132 randomly selected representatives of 92 other breeds. Although the PIGN:c.398T allele was not present in the Poodles that we genotyped, it could still be carried by other Poodles. Nonetheless, an alternative and more likely explanation for the origin of the three homozygous Whoodles is that they resulted from a Whoodle to SCWT backcross or from two Whoodle parents.
Flow cytometry
The biological activities of mutant PIGN isoforms were assessed by transiently transfecting corresponding cDNAs into PIGN-knockout HEK293 cells and measuring the restoration of GPI-anchored CD59 surface expression [38, 39]. Flow cytometry of cells which were transfected with a medium promoter-driven expression vector revealed comparable restoration of CD59 cell-surface expression by PIGN-knockout HEK293 cells 3 days after transient transfection with wild-type human PIGN cDNA or with recombinant human cDNAs designed to encode PIGN:p. T133S or PIGN:p. T133V isoforms. However, transient transfection with recombinant human cDNAs designed to encode PIGN:p. T133I restored only 60–70% of the cell-surface CD59 that was produced by the wild-type cDNA (Fig. 2a). The same T331I PIGN cDNA, when expressed with a strong promoter-driven vector pME, nearly fully restored CD59 (Fig. 2a). Western blotting revealed comparably expressed protein levels of T331I and wild-type PIGN (Fig. 2b). It is therefore concluded that the T331I mutation causes a reduction in specific activity rather than stability of PIGN.
Functional analysis of PIGN mutants in PIGN-knockout HEK293 cells. a Restoration of cell-surface expression of GPI-anchored protein CD59 on PIGN-knockout HEK293 cells after transfection. HA-epitope-tagged, wild-type T331I, T331S, and T331V human PIGN cDNAs were transfected with a medium promoter-driven pTK vector (top) or a strong promoter-driven pME vector (bottom). Three days later, CD59 levels were assessed by flow cytometry. b Western blotting analysis of wild-type T331I, T331S, and T331V human PIGN. PIGN-knockout HEK293 cells that were transfected with HA-epitope-tagged, wild-type and mutant PIGN cDNAs in pME were analyzed 3 days later by western blotting using anti-HA antibody. GAPDH glyceraldehyde 3-phosphate dehydrogenase, a loading control
Discussion
We studied a cPxD originally described in SCWT in 2004 [31], which shared features of human PxD. All the affected dogs were normal on examination between episodes. The movements appeared to be involuntary and could not be disrupted. The dogs remained conscious during the episodes, and except for severe episodes, the dogs continued to walk, suggesting that the movements were semi-purposeful. Some of the movements in this cPxD were the sustained, abnormal postures of a dystonia [40]. The more rapid movements in cPxD are hyperkinetic movements, but they are difficult to place into one of the human categories of chorea, ballism, or athetosis [41]. The various forms of human PxD can be subclassified as a PED, PKD, or PNKD [1–4, 6, 7]. The owners of dogs with cPxD never mentioned exercise or fasting as triggers of dyskinesia episodes as occurs with PED in humans [42]. Determining whether a sudden movement triggered the episodes as in PKD in humans [7] was difficult since the owners typically were not closely observing their dogs at the onset of the episodes. Compared to the durations and frequencies of episodes of dyskinesia in human PKD patients [7], the episodes in dogs with PIGN-associated cPxD lasted longer (minutes to hours) and occurred less frequently (several/day to every few weeks), more like PNKD in humans [6]. Stress and excitement, which trigger episodes in most PNKD associated with PNKD in humans [6], were reported to trigger the episodes in some dogs, although in most cases, no trigger could be identified. While alcohol or caffeine ingestion are the most consistent triggers of human PNKD associated with PNKD mutations [6], dogs rarely ingest alcohol and methylxanthines are toxic to dogs [43], so whether they would trigger episodes in dogs is unknown. PKD patients either require no treatment or respond to anti-epileptic drugs, and PNKD patients often respond to benzodiazepines [2, 5–7], while SCWT with cPxD showed a poor response to these drugs. The median age of onset in dogs was 2.25 years, a young but fully mature adult dog, which would be older than the equivalent average age of onset in humans of PNKD associated with PNKD or PKD and more comparable to “atypical” PNKD not associated with PNKD mutations [6, 7]. Thus, even though there are differences, the dyskinesia episodes in PIGN-associated cPxD most closely resemble those of human PNKD patients.
To identify the molecular genetic cause of cPxD, we generated whole genome sequences for two affected dogs. The T allele of a PIGN:c.398C > T transition was the only rare, homozygous, predicted amino acid altering sequence variant that was found in both of these animals. All 25 of our c.398T allele homozygous DNA samples were from cPxD-affected dogs, whereas none of the 1053 control SCWT and Poodles and none of the 132 genotyped dogs from other breeds were c.398T homozygotes. This established a strong association between the clinical disease and T allele homozygosity. None of the 15 PIGN:c.398C/T heterozygous SCWT were known to have exhibited episodes of dyskinesia, which supports the inheritance of SCWT cPxD as an autosomal recessive trait.
PIGN is 1 of more than 26 genes that encode the polypeptides which contribute to the biosynthesis, protein attachment, and remodeling of GPI anchors. GPI anchors tether a diverse group of cell-surface proteins to the plasma membrane (Fig. 3a). Amide bonds covalently attach the cell-surface protein to an ethanolamine phosphate at the outer end of the GPI anchor, while at the inner end, aliphatic chains intercalate into the plasma membranes and bind to hydrophobic lipid-raft components. A complex core glycolipid provides a covalent bridge between the two ends of the GPI anchor [39, 44]. During GPI assembly in the endoplasmic reticulum, ethanolamine phosphate side chains are added to mannosyl moieties in the core glycolipid. GPI ethanolamine phosphate transferase-1 (PIGN), the enzyme encoded by PIGN, facilitates the transfer of an ethanolamine phosphate from phosphatidylethanolamine to the innermost mannosyl moiety in a partially assembled core glycolipid intermediate (Fig. 3b) [45–47]. The existence of PIGN-associated deficiency diseases indicates that this ethanolamine phosphate side chain has an important biological function, probably related to its role in the recognition of GPI glycolipids by the transamidase complex that attaches proteins to the GPI anchors [48]. Flow cytometry has been used to demonstrate altered expression of these proteins with mutations of PIGN in humans [38, 49–51].
Structure and synthesis of GPI. a The core structure of the GPI anchor is a phosphatidylinositol moiety, a glucosamine moiety, three mannoses, and an ethanolamine phosphate (EtNP) on the terminal mannose. The lipid tails of PI are embedded in lipid rafts in the plasma membrane, and cell-surface proteins are bound to the terminal EtNP. Another EtNP bound to the first mannose is found in all mammalian cells. b PIGN adds the EtNP to the first mannose during the synthesis of GPI in the endoplasmic reticulum [39]
The variant PIGN:c.398T allele is predicted to result in a missense mutation, PIGN:p. T133I, in the phosphodiesterase domain. Evidence that the PIGN:p. T133I substitution partially impairs biological function was obtained by transiently transfecting PIGN-knockout HEK293 cells with a human PIGN cDNA construct designed to encode the p. T133I substitution. As demonstrated by flow cytometry, the human PIGN:p. T133I cDNA was ∼60% as effective as wild-type PIGN cDNA in restoring CD59 surface antigen to the PIGN-knockout cells. Available sequence information indicates that PIGN orthologs in at least 146 vertebrate species have a threonine codon at the position orthologous to codon 133 in the human and canine PIGN; however, at least two vertebrate species, the pig and the cape elephant shrew, have a serine codon at this position (Supplementary Table 2). This suggested that the hydroxyl moiety of threonine or serine might be required for normal biological function. To test this hypothesis, we transiently transfected PIGN-knockout HEK293 cells with recombinant human PIGN cDNA designed to encode either PIGN:p. T133S or PIGN:p. T133V. With both of these constructs, the restoration of CD59 surface antigen was similar to that produced by the wild-type PIGN cDNA. The apparently normal function of PIGN with a valine at position 133 indicates that hydroxyl moiety at this position is not a functional requirement. Nonetheless, the contrast in flow cytometry patterns obtained with the various cDNAs substantiates the significance of the 30–40% decrease in CD59 surface antigen restoration that was obtained with the cDNA containing the p. T133I variant.
N-ethyl-N-nitrosourea-derived mice with a homozygous truncating splice-site mutation in Pign die in utero with developmental deficiencies including holoprosencephaly [52]. Similarly, a human fetus died with a diaphragmatic hernia and many other severe developmental abnormalities, apparently due to a homozygous nullifying splice-site mutation [53]. Individuals born with other homozygous or compound heterozygous PIGN mutations share a developmental deficiency syndrome known as multiple congenital anomalies-hypotonia-seizures syndrome-1 or MCAHS1 (OMIM #614,080). The current literature describes at least 17 MCAHS1 patients from 9 families carrying 13 different PIGN mutations [38, 49–51, 54, 55]. These patients had neonatal hypotonia, severe developmental delays, congenital anomalies, visual impairment, hyporeflexia, tremors, and seizures. Some exhibited choreoathetosis and gait abnormalities, while others had structural brain abnormalities including delayed myelination, cortical atrophy, diffusion restriction in the globus pallidi and corticospinal tracts, and cerebellar atrophy or parenchymal loss in the vermis [38, 51, 55]. The severity of clinical signs in MCAHS1 patients appears to correlate with the functional severity of the PIGN mutations. The most severe signs, which included multiple structural and functional developmental anomalies, occurred in a neonate with a frameshift mutation in trans with a multi-exon microdeletion. In contrast, a child with likely hypomorphic compound heterozygous missense PIGN mutations showed no dysmorphic features, moderate developmental delay, and a later onset of seizures and spastic quadriparesis [55].
The cPxD described in the current report is less severe than the human and murine PIGN-associated phenotypes. In contrast to the developmental delays and early mortality seen in humans [38, 51, 55] and mice [52], respectively, with loss of PIGN function, the first episodes of dyskinesia occurred in young-adult dogs. The dogs appeared to be normal prior to the onset of the dyskinesia and between episodes. The marked differences in severity may be due to inherent differences between species. Alternatively, the differences in severity may occur because the canine mutation encodes a product which retains a relatively greater amount of residual functional activity. This second possibility is supported by the finding that transfection of PIGN-knockout HEK293 cells with human T133I PIGN cDNA in a strong promoter-driven pME vector fully restored CD59 surface expression, whereas a recent report [38] showed that transfection of PIGN-knockout HEK293 cells with the same strong promoter-driven vector and PIGN cDNA carrying the MCAHS1-associated S270P resulted in markedly reduced CD59 surface expression (see Fig. 2b in reference [38]). Thus, it seems possible that hypomorphic mutations of human PIGN could also cause PxD or other disease phenotypes that are less severe than those of MCAHS1. Similar variability in phenotype is seen in SLC2A1 variants associated with PxD in humans [14, 15, 56].
In summary, we have described a paroxysmal dyskinesia in dogs that most closely resembles PNKD but differs from the known human PxDs; in that, it is autosomal recessively inherited. This disease is very likely caused by a hypomorphic missense mutation in PIGN, which encodes an enzyme in the biosynthetic pathway for GPI anchors. If so, it expands the phenotypes associated with altered GPI function and suggests candidate genes to investigate in human PxD that have not had a causal mutation identified. The spontaneous canine disease could serve as an animal model to investigate the pathogenesis of PxD and potential therapies. Nonetheless, we cannot rule out the possibility that a variant in tight linkage disequilibrium with the PIGN:c.398C > T transition is the true cause of the disease. Generation of a mouse model expressing the PIGN:c.398T variant that recapitulated the phenotype seen in dogs or the identification of other PIGN variants in dogs with cPxD would help confirm that the variant is the causal and permit further studies on its effect on GPI function and the difference in phenotype with the null mutations in mice and humans.
References
Demirkiran M, Jankovic J (1995) Paroxysmal dyskinesias: clinical features and classification. Ann Neurol 38(4):571–579. doi:10.1002/ana.410380405
Bhatia KP (2011) Paroxysmal dyskinesias. Mov Disord 26(6):1157–1165
Jankovic J, Demirkiran M (2002) Classification of paroxysmal dyskinesias and ataxias. In: Fahn S, Frucht SJ, Hallett M, Truong DD (eds) Myoclonus and paroxysmal dyskinesia, vol 89. Advances in neurology. Lippincott Williams & Wilkins, Philadelphia, pp. 387–400
Waln O, Jankovic J (2015) Paroxysmal movement disorders. Neurol Clin 33(1):137–152
Erro R, Sheerin UM, Bhatia KP (2014) Paroxysmal dyskinesias revisited: a review of 500 genetically proven cases and a new classification. Mov Disord 29(9):1108–1116. doi:10.1002/mds.25933
Bruno MK, Lee HY, Auburger GW, Friedman A, Nielsen JE, Lang AE, Bertini E, Van Bogaert P, Averyanov Y, Hallett M, Gwinn-Hardy K, Sorenson B, Pandolfo M, Kwiecinski H, Servidei S, Fu YH, Ptacek L (2007) Genotype-phenotype correlation of paroxysmal nonkinesigenic dyskinesia. Neurology 68(21):1782–1789. doi:10.1212/01.wnl.0000262029.91552.e0
Bruno MK, Hallett M, Gwinn-Hardy K, Sorensen B, Considine E, Tucker S, Lynch DR, Mathews KD, Swoboda KJ, Harris J, Soong BW, Ashizawa T, Jankovic J, Renner D, Fu YH, Ptacek LJ (2004) Clinical evaluation of idiopathic paroxysmal kinesigenic dyskinesia: new diagnostic criteria. Neurology 63(12):2280–2287
Blakeley J, Jankovic J (2002) Secondary paroxysmal dyskinesias. Mov Disord: Off J Mov Disord Soc 17(4):726–734. doi:10.1002/mds.10178
Chen WJ, Lin Y, Xiong ZQ, Wei W, Ni W, Tan GH, Guo SL, He J, Chen YF, Zhang QJ, Li HF, Lin Y, Murong SX, Xu J, Wang N, Wu ZY (2011) Exome sequencing identifies truncating mutations in PRRT2 that cause paroxysmal kinesigenic dyskinesia. Nat Genet 43(12):1252–1255. doi:10.1038/ng.1008
Rainier S, Thomas D, Tokarz D, Ming L, Bui M, Plein E, Zhao X, Lemons R, Albin R, Delaney C, Alvarado D, Fink JK (2004) Myofibrillogenesis regulator 1 gene mutations cause paroxysmal dystonic choreoathetosis. Arch Neurol 61(7):1025–1029. doi:10.1001/archneur.61.7.1025
Suls A, Dedeken P, Goffin K, Van EH, Dupont P, Cassiman D, Kempfle J, Wuttke TV, Weber Y, Lerche H, Afawi Z, Vandenberghe W, Korczyn AD, Berkovic SF, Ekstein D, Kivity S, Ryvlin P, Claes LR, Deprez L, Maljevic S, Vargas A, Van DT, Goossens D, Del-Favero J, Van LK, De JP, Van PW (2008) Paroxysmal exercise-induced dyskinesia and epilepsy is due to mutations in SLC2A1, encoding the glucose transporter GLUT1. Brain 131(Pt:7):1831–1844
Wang JL, Cao L, Li XH, Hu ZM, Li JD, Zhang JG, Liang Y, San A, Li N, Chen SQ, Guo JF, Jiang H, Shen L, Zheng L, Mao X, Yan WQ, Zhou Y, Shi YT, Ai SX, Dai MZ, Zhang P, Xia K, Chen SD, Tang BS (2011) Identification of PRRT2 as the causative gene of paroxysmal kinesigenic dyskinesias. Brain 134(Pt 12):3493–3501. doi:10.1093/brain/awr289
Lee HY, Xu Y, Huang Y, Ahn AH, Auburger GW, Pandolfo M, Kwiecinski H, Grimes DA, Lang AE, Nielsen JE, Averyanov Y, Servidei S, Friedman A, Van Bogaert P, Abramowicz MJ, Bruno MK, Sorensen BF, Tang L, Fu YH, Ptacek LJ (2004) The gene for paroxysmal non-kinesigenic dyskinesia encodes an enzyme in a stress response pathway. Hum Mol Genet 13(24):3161–3170. doi:10.1093/hmg/ddh330
Brockmann K (2009) The expanding phenotype of GLUT1-deficiency syndrome. Brain and Development 31(7):545–552. doi:10.1016/j.braindev.2009.02.008
Anand G, Padeniya A, Hanrahan D, Scheffer H, Zaiwalla Z, Cox D, Mann N, Hewertson J, Price S, Nemeth A, Arsov T, Scheffer I, Jayawant S, Pike M, McShane T (2011) Milder phenotypes of glucose transporter type 1 deficiency syndrome. Dev Med Child Neurol 53(7):664–668. doi:10.1111/j.1469-8749.2011.03949.x
Schneider SA, Paisan-Ruiz C, Garcia-Gorostiaga I, Quinn NP, Weber YG, Lerche H, Hardy J, Bhatia KP (2009) GLUT1 gene mutations cause sporadic paroxysmal exercise-induced dyskinesias. Mov Disord 24(11):1684–1688. doi:10.1002/mds.22507
Weber YG, Storch A, Wuttke TV, Brockmann K, Kempfle J, Maljevic S, Margari L, Kamm C, Schneider SA, Huber SM, Pekrun A, Roebling R, Seebohm G, Koka S, Lang C, Kraft E, Blazevic D, Salvo-Vargas A, Fauler M, Mottaghy FM, Munchau A, Edwards MJ, Presicci A, Margari F, Gasser T, Lang F, Bhatia KP, Lehmann-Horn F, Lerche H (2008) GLUT1 mutations are a cause of paroxysmal exertion-induced dyskinesias and induce hemolytic anemia by a cation leak. J Clin Invest 118(6):2157–2168. doi:10.1172/jci34438
Li M, Niu F, Zhu X, Wu X, Shen N, Peng X, Liu Y (2015) PRRT2 mutant leads to dysfunction of glutamate signaling. Int J Mol Sci 16(5):9134–9151. doi:10.3390/ijms16059134
Nobile C, Striano P (2014) PRRT2: a major cause of infantile epilepsy and other paroxysmal disorders of childhood. Prog Brain Res 213:141–158. doi:10.1016/b978-0-444-63326-2.00008-9
Shen Y, Ge WP, Li Y, Hirano A, Lee HY, Rohlmann A, Missler M, Tsien RW, Jan LY, Fu YH, Ptacek LJ (2015) Protein mutated in paroxysmal dyskinesia interacts with the active zone protein RIM and suppresses synaptic vesicle exocytosis. Proc Natl Acad Sci U S A 112(10):2935–2941. doi:10.1073/pnas.1501364112
Du W, Bautista JF, Yang H, Diez-Sampedro A, You SA, Wang L, Kotagal P, Luders HO, Shi J, Cui J, Richerson GB, Wang QK (2005) Calcium-sensitive potassium channelopathy in human epilepsy and paroxysmal movement disorder. Nat Genet 37(7):733–738. doi:10.1038/ng1585
Castiglioni C, Verrigni D, Okuma C, Diaz A, Alvarez K, Rizza T, Carrozzo R, Bertini E, Miranda M (2015) Pyruvate dehydrogenase deficiency presenting as isolated paroxysmal exercise induced dystonia successfully reversed with thiamine supplementation. Case report and mini-review. Eur J Paediatr Neurol 19(5):497–503. doi:10.1016/j.ejpn.2015.04.008
Urkasemsin G, Olby N (2014) Canine paroxysmal movement disorders. Vet Clin North Am Small Anim Pract 44(6):1091–1102
Penderis J, Franklin RJ (2001) Dyskinesia in an adult bichon frise. J Small Anim Pract 42(1):24–25
Black V, Garosi L, Lowrie M, Harvey RJ, Gale J (2014) Phenotypic characterisation of canine epileptoid cramping syndrome in the border terrier. J Small Anim Pract 55:102–107. doi:10.1111/jsap.12170
Herrtage ME, Palmer AC (1983) Episodic falling in the cavalier king Charles spaniel. Vet Rec 112(19):458–459
Packer RA, Patterson EE, Taylor JF, Coates JR, Schnabel RD, O’Brien DP (2010) Characterization and mode of inheritance of a paroxysmal dyskinesia in Chinook dogs. J Vet Intern Med 24(6):1305–1313
Wolf M, Bruehschwein A, Sauter-Louis C, Sewell AC, Fischer A (2011) An inherited episodic head tremor syndrome in Doberman pinscher dogs. Mov Disord 26(13):2381–2386
Guevar J, De Decker S, Van Ham LM, Fischer A, Volk HA (2014) Idiopathic head tremor in English bulldogs. Mov Disord 29(2):191–194. doi:10.1002/mds.25767
Meyers KM, Lund JE, Padgett G, Dickson WM (1969) Hyperkinetic episodes in Scottish terrier dogs. J Am Vet Med Assoc 155(2):129–133
Shelton GD (2004) Muscle pain, cramps and hypertonicity. Vet Clin North Am Small Anim Pract 34(6):1483–1496. doi:10.1016/j.cvsm.2004.05.019
Forman OP, Penderis J, Hartley C, Hayward LJ, Ricketts SL, Mellersh CS (2012) Parallel mapping and simultaneous sequencing reveals deletions in BCAN and FAM83H associated with discrete inherited disorders in a domestic dog breed. PLoS Genet 8(1):e1002462. doi:10.1371/journal.pgen.1002462
Gill JL, Tsai KL, Krey C, Noorai RE, Vanbellinghen JF, Garosi LS, Shelton GD, Clark LA, Harvey RJ (2012) A canine BCAN microdeletion associated with episodic falling syndrome. NeurobiolDis 45(1):130–136
Gilliam D, O’Brien DP, Coates JR, Johnson GS, Johnson GC, Mhlanga-Mutangadura T, Hansen L, Taylor JF, Schnabel RD (2014) A homozygous KCNJ10 mutation in Jack Russell terriers and related breeds with spinocerebellar ataxia with myokymia, seizures, or both. J Vet Intern Med 28(3):871–877. doi:10.1111/jvim.12355
Guo J, O’Brien DP, Mhlanga-Mutangadura T, Olby NJ, Taylor JF, Schnabel RD, Katz ML, Johnson GS (2015) A rare homozygous MFSD8 single-base-pair deletion and frameshift in the whole genome sequence of a Chinese crested dog with neuronal ceroid lipofuscinosis. BMC Vet Res 10(1):960. doi:10.1186/s12917-014-0181-z
Guo J, Johnson GS, Brown HA, Provencher ML, da Costa RC, Mhlanga-Mutangadura T, Taylor JF, Schnabel RD, O’Brien DP, Katz ML (2014) A CLN8 nonsense mutation in the whole genome sequence of a mixed breed dog with neuronal ceroid lipofuscinosis and Australian shepherd ancestry. Mol Genet Metab 112(4):302–309. doi:10.1016/j.ymgme.2014.05.014
Livak KJ (1999) Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet Anal 14(5–6):143–149
Ohba C, Okamoto N, Murakami Y, Suzuki Y, Tsurusaki Y, Nakashima M, Miyake N, Tanaka F, Kinoshita T, Matsumoto N, Saitsu H (2013) PIGN mutations cause congenital anomalies, developmental delay, hypotonia, epilepsy, and progressive cerebellar atrophy. Neurogenetics 15(2):85–92. doi:10.1007/s10048-013-0384-7
Kinoshita T (2014) Biosynthesis and deficiencies of glycosylphosphatidylinositol. Proc Jpn Acad Ser B, Phys Biol Sci 90(4):130–143
Sanger TD, Delgado MR, Gaebler-Spira D, Hallett M, Mink JW (2003) Classification and definition of disorders causing hypertonia in childhood. Pediatrics 111(1):e89–e97
Sanger TD, Chen D, Fehlings DL, Hallett M, Lang AE, Mink JW, Singer HS, Alter K, Ben-Pazi H, Butler EE, Chen R, Collins A, Dayanidhi S, Forssberg H, Fowler E, Gilbert DL, Gorman SL, Gormley ME Jr, Jinnah HA, Kornblau B, Krosschell KJ, Lehman RK, MacKinnon C, Malanga CJ, Mesterman R, Michaels MB, Pearson TS, Rose J, Russman BS, Sternad D, Swoboda KJ, Valero-Cuevas F (2010) Definition and classification of hyperkinetic movements in childhood. Mov Disord 25(11):1538–1549
Bhatia KP, Soland VL, Bhatt MH, Quinn NP, Marsden CD (1997) Paroxysmal exercise-induced dystonia: eight new sporadic cases and a review of the literature. Mov Disord: Off J Mov Disord Soc 12(6):1007–1012. doi:10.1002/mds.870120626
Hooser SB, Beasley VR (1986) Methylxanthine poisoning (chocolate and caffeine toxicosis). In: Kirk RW (ed) Current veterinary therapy IX. WB Saudners, Philadelphia, pp. 191–192
Kinoshita T, Fujita M, Maeda Y (2008) Biosynthesis, remodelling and functions of mammalian GPI-anchored proteins: recent progress. J Biochem 144(3):287–294. doi:10.1093/jb/mvn090
Orlean P, Menon AK (2007) Thematic review series: lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. J Lipid Res 48(5):993–1011. doi:10.1194/jlr.R700002-JLR200
Gaynor EC, Mondesert G, Grimme SJ, Reed SI, Orlean P, Emr SD (1999) MCD4 encodes a conserved endoplasmic reticulum membrane protein essential for glycosylphosphatidylinositol anchor synthesis in yeast. Mol Biol Cell 10(3):627–648
Hong Y, Maeda Y, Watanabe R, Ohishi K, Mishkind M, Riezman H, Kinoshita T (1999) Pig-n, a mammalian homologue of yeast Mcd4p, is involved in transferring phosphoethanolamine to the first mannose of the glycosylphosphatidylinositol. J Biol Chem 274(49):35099–35106
Vainauskas S, Menon AK (2006) Ethanolamine phosphate linked to the first mannose residue of glycosylphosphatidylinositol (GPI) lipids is a major feature of the GPI structure that is recognized by human GPI transamidase. J Biol Chem 281(50):38358–38364. doi:10.1074/jbc.M608896200
Khayat M, Tilghman JM, Chervinsky I, Zalman L, Chakravarti A, Shalev SA (2015) A PIGN mutation responsible for multiple congenital anomalies-hypotonia-seizures syndrome 1 (MCAHS1) in an Israeli-Arab family. Am J Med Genet A 170(1):176–182. doi:10.1002/ajmg.a.37375
Nakagawa T, Taniguchi-Ikeda M, Murakami Y, Nakamura S, Motooka D, Emoto T, Satake W, Nishiyama M, Toyoshima D, Morisada N, Takada S, Tairaku S, Okamoto N, Morioka I, Kurahashi H, Toda T, Kinoshita T, Iijima K (2016) A novel PIGN mutation and prenatal diagnosis of inherited glycosylphosphatidylinositol deficiency. Am J Med Genet A 170(1):183–188. doi:10.1002/ajmg.a.37397
Maydan G, Noyman I, Har-Zahav A, Neriah ZB, Pasmanik-Chor M, Yeheskel A, Albin-Kaplanski A, Maya I, Magal N, Birk E, Simon AJ, Halevy A, Rechavi G, Shohat M, Straussberg R, Basel-Vanagaite L (2011) Multiple congenital anomalies-hypotonia-seizures syndrome is caused by a mutation in PIGN. J Med Genet 48(6):383–389. doi:10.1136/jmg.2010.087114
McKean DM, Niswander L (2012) Defects in GPI biosynthesis perturb Cripto signaling during forebrain development in two new mouse models of holoprosencephaly. Biology open 1(9):874–883. doi:10.1242/bio.20121982
Brady PD, Moerman P, De Catte L, Deprest J, Devriendt K, Vermeesch JR (2014) Exome sequencing identifies a recessive PIGN splice site mutation as a cause of syndromic congenital diaphragmatic hernia. Eur J Med Genet 57(9):487–493. doi:10.1016/j.ejmg.2014.05.001
Couser NL, Masood MM, Strande NT, Foreman AK, Crooks K, Weck KE, Lu M, Wilhelmsen KC, Roche M, Evans JP, Berg JS, Powell CM (2015) The phenotype of multiple congenital anomalies-hypotonia-seizures syndrome 1: report and review. Am J Med Genet A 167A(9):2176–2181. doi:10.1002/ajmg.a.37129
Fleming L, Lemmon M, Beck N, Johnson M, Mu W, Murdock D, Bodurtha J, Hoover-Fong J, Cohn R, Bosemani T, Baranano K, Hamosh A (2015) Genotype-phenotype correlation of congenital anomalies in multiple congenital anomalies hypotonia seizures syndrome (MCAHS1)/PIGN-related epilepsy. Am J Med Genet A 170(1):77–86. doi:10.1002/ajmg.a.37369
Groffen AJ, Klapwijk T, van Rootselaar AF, Groen JL, Tijssen MA (2013) Genetic and phenotypic heterogeneity in sporadic and familial forms of paroxysmal dyskinesia. J Neurol 260(1):93–99. doi:10.1007/s00415-012-6592-5
Acknowledgments
We thank all the pet owners, breeders, and veterinarians who helped with the data collection and provided videos especially Rebecca Packer, Stephanie Thomovsky, Jason Berg, and Laura Vasquez. We also thank Robert Wayne of the University of California Los Angeles, Matt Huntelman of the Translational Genomics Research Institute, Kate Meurs and Josh Stern of North Carolina State University, and Paula Henthorn of the University of Pennsylvania for providing the genome sequence data for use as controls. We thank Dr. Tomohiro Chiyonobu, Kyoto Prefectural University of Medicine, for the helpful discussion.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All studies were approved by the University of Missouri Animal Care and Use Committee, and informed consent was obtained from the dogs’ owners.
Electronic supplementary material
Table S1
(DOCX 44 kb)
Table S2
(GIF 3996 kb)
High Resolution Table S2
(TIFF 39334 kb)
Video S1
(WMV 5762 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kolicheski, A.L., Johnson, G.S., Mhlanga-Mutangadura, T. et al. A homozygous PIGN missense mutation in Soft-Coated Wheaten Terriers with a canine paroxysmal dyskinesia. Neurogenetics 18, 39–47 (2017). https://doi.org/10.1007/s10048-016-0502-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-016-0502-4