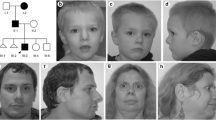
Abstract
We report a girl with intellectual disability (ID), neuropsychiatric alterations, and a de novo balanced t(10;19)(q22.3;q13.33) translocation. After chromosome sorting, fine mapping of breakpoints by array painting disclosed disruptions of the zinc finger, MIZ-type containing 1 (ZMIZ1) (on chr10) and proline-rich 12 (PRR12) (on chr19) genes. cDNA analyses revealed that the translocation resulted in gene fusions. The resulting hybrid transcripts predict mRNA decay or, if translated, formation of truncated proteins, both due to frameshifts that introduced premature stop codons. Though other molecular mechanisms may be operating, these results suggest that haploinsufficiency of one or both genes accounts for the patient’s phenotype. ZMIZ1 is highly expressed in the brain, and its protein product appears to interact with neuron-specific chromatin remodeling complex (nBAF) and activator protein 1 (AP-1) complexes which play a role regulating the activity of genes essential for normal synapse and dendrite growth/behavior. Strikingly, the patient’s phenotype overlaps with phenotypes caused by mutations in SMARCA4 (BRG1), an nBAF subunit presumably interacting with ZMIZ1 in brain cells as suggested by our results of coimmunoprecipitation in the mouse brain. PRR12 is also expressed in the brain, and its protein product possesses domains and residues thought to be related in formation of large protein complexes and chromatin remodeling. Our observation from E15 mouse brain cells that a Prr12 isoform was confined to nucleus suggests a role as a transcription nuclear cofactor likely involved in neuronal development. Moreover, a pilot transcriptome analysis from t(10;19) lymphoblastoid cell line suggests dysregulation of genes linked to neurodevelopment processes/neuronal communication (e.g., NRCAM) most likely induced by altered PRR12. This case represents the first constitutional balanced translocation disrupting and fusing both genes and provides clues for the potential function and effects of these in the central nervous system.





Similar content being viewed by others
References
Bugge M, Bruun-Petersen G, Brøndum-Nielsen K et al (2000) Disease associated balanced chromosome rearrangements: a resource for large scale genotype-phenotype delineation in man. J Med Genet 37:858–865
Yue Y, Grossmann B, Holder SE, Haaf T (2005) De novo t(7;10)(q33;q23) translocation and closely juxtaposed microdeletion in a patient with macrocephaly and developmental delay. Hum Genet 117:1–8
De Gregori M, Ciccone R, Magini P et al (2007) Cryptic deletions are a common finding in “balanced” reciprocal and complex chromosome rearrangements: a study of 59 patients. J Med Genet 44:750–762
Stankiewicz P, Beaudet A (2007) Use of array CGH in the evaluation of dysmorphology, malformations, developmental delay, and idiopathic mental retardation. Curr Opin Genet Dev 17:182–192
Borsani G, Piovani G, Zoppi N et al (2008) Cytogenetic and molecular characterization of a de-novo t(2p;7p) translocation involving TNS3 and EXOC6B genes in a boy with a complex syndromic phenotype. Eur J Med Genet 51:292–302
Schluth-Bolard C, Delobel B, Sanlaville D et al (2009) Cryptic genomic imbalances in de novo and inherited apparently balanced chromosomal rearrangements: array CGH study of 47 unrelated cases. Eur J Med Genet 52:291–296
Vandeweyer G, Kooy RF (2009) Balanced translocations in mental retardation. Hum Genet 126:133–147
Backx L, Seuntjens E, Devriendt K et al (2011) A balanced translocation t(6;14)(q25.3;q13.2) leading to reciprocal fusion transcripts in a patient with intellectual disability and agenesis of corpus callosum. Cytogenet Genome Res 132:135–143
Ropers HH (2007) New perspectives for the elucidation of genetic disorders. Am J Hum Genet 81:199–207
Laumonnier F, Cuthbert PC, Grant SG (2007) The role of neuronal complexes in human X-linked brain diseases. Am J Hum Genet 80:205–220
Tsurusaki Y, Okamoto N, Ohashi H et al (2012) Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat Genet 44:376–378
Kim HG, Kim HT, Leach NT et al (2012) Translocations disrupting PHF21A in the Potocki-Shaffer-syndrome region are associated with intellectual disability and craniofacial anomalies. Am J Hum Genet 91:56–72
Fiegler H, Gribble SM, Burford DC et al (2003) Array painting: a method for the rapid analysis of aberrant chromosomes using DNA microarrays. J Med Genet 40:664–670
Veltman IM, Veltman JA, Arkesteijn G et al (2003) Chromosomal breakpoint mapping by arrayCGH using flow-sorted chromosomes. Biotechniques 35:1066–1070
Arkesteijn G, Jumelet E, Hagenbeek A et al (1999) Reverse chromosome painting for the identification of marker chromosomes and complex translocations in leukemia. Cytometry 35:117–124
Chen W, Erdogan F, Ropers HH et al (2005) CGHPRO—a comprehensive data analysis tool for array CGH. BMC Bioinforma 6:85
Carlin RK, Grab DJ, Cohen RS, Siekevitz P (1980) Isolation and characterization of postsynaptic densities from various brain regions: enrichment of different types of postsynaptic densities. J Cell Biol 86:831–845
Lovtrup-Rein H, McEwen BS (1966) Isolation and fractionation of rat brain nuclei. J Cell Biol 30:405–415
Delint-Ramírez I, Salcedo-Tello P, Bermudez-Rattoni F (2008) Spatial memory formation induces recruitment of NMDA receptor and PSD-95 to synaptic lipid rafts. J Neurochem 106:1658–1668
Bolotin E, Armendariz A, Kim K et al (2014) Statin-induced changes in gene expression in EBV-transformed and native B-cells. Hum Mol Genet 23:1202–1210
Jensen P, Magdaleno S, Lehman KM et al (2004) Aneurogenomics approach to gene expression analysis in the developing brain. Brain Res Mol Brain Res 132:116–127
Sharma M, Li X, Wang Y et al (2003) hZimp10 is an androgen receptor co-activator and forms a complex with SUMO-1 at replication foci. EMBO J 22:6101–6114
Beliakoff J, Lee J, Ueno H et al (2008) The PIAS-like protein Zimp10 is essential for embryonic viability and proper vascular development. Mol Cell Biol 28:282–292
Henderson P, van Limbergen JE, Wilson DC et al (2011) Genetics of childhood-onset inflammatory bowel disease. Inflamm Bowel Dis 17:346–361
Rodriguez-Magadán H, Merino E, Schnabel D et al (2008) Spatial and temporal expression of Zimp7 and Zimp10 PIAS-like proteins in the developing mouse embryo. Gene Expr Patterns 8:206–213
Nagase T, Ishikawa K, Kikuno R et al (1999) Prediction of the coding sequences of unidentified human genes. XV. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res 6:337–345
Darnell JC, Van Driesche SJ, Zhang C et al (2011) FMRP Stalls Ribosomal Translocation on mRNAs Linked to Synaptic Function and Autism. Cell 146:247–261
Li X, Zhu C, Tu WH et al (2011) ZMIZ1 preferably enhances the transcriptional activity of androgen receptor with short polyglutamine tract. PLoS One 6, e25040
Wu JI, Lessard J, Olave IA et al (2007) Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron 56:94–108
Gass P, Fleischmann A, Hvalby O et al (2004) Mice with a fra-1 knock-in into the c-fos locus show impaired spatial but regular contextual learning and normal LTP. Brain Res Mol Brain Res 130:16–22
Alberini CM (2009) Transcription factors in long-term memory and synaptic plasticity. Physiol Rev 89:121–145
Pérez-Cadahía B, Drobic B, Davie JR (2011) Activation and function of immediate-early genes in the nervous system. Biochem Cell Biol 89:61–73
Vonhoff F, Kuehn C, Blumenstock S et al (2013) Temporal coherency between receptor expression, neural activity and AP-1-dependent transcription regulates Drosophila motoneuron dendrite development. Development 140:606–616
Ferreira MA, O'Donovan MC, Meng YA et al (2008) Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet 40:1056–1058
Disanto G, Sandve GK, Berlanga-Taylor AJ et al (2012) Vitamin D receptor binding, chromatin states and association with multiple sclerosis. Hum Mol Genet 21:3575–3586
Amunts K, Kedo O, Kindler M et al (2005) Cytoarchitectonic mapping of the human amygdala, hippocampal region and entorhinal cortex: intersubject variability and probability maps. Anat Embryol (Berl) 210:343–352
Hussain R, Ghoumari AM, Bielecki B et al (2013) The neural androgen receptor: a therapeutic target for myelin repair in chronic demyelination. Brain 136:132–146
Cunningham RL, Lumia AR, McGinnis MY (2012) Androgen receptors, sex behavior, and aggression. Neuroendocrinology 96:131–140
Kosho T, Okamoto N, Ohashi H et al (2013) Clinical Correlations of Mutations Affecting Six Components of the SWI/SNF Complex: Detailed Description of 21 Patients and a Review of the Literature. Am J Med Genet A 161A:1221–1237
Rigbolt KT, Prokhorova TA, Akimov V et al (2011) System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signal 4(164):rs3
Aravind L, Landsman D (1998) AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res 26:4413–4421
Choudhary C, Kumar C, Gnad F et al (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325:834–840
Baker SA, Chen L, Wilkins AD et al (2013) An AT-hook domain in MeCP2 determines the clinical course of Rett syndrome and related disorders. Cell 152:984–996
Murata Y, Doi T, Taniguchi H, Fujiyoshi Y (2005) Proteomic analysis revealed a novel synaptic proline-rich membrane protein (PRR7) associated with PSD-95 and NMDA receptor. Biochem Biophys Res Commun 327:183–191
Sakurai T (2012) The role of NrCAM in neural development and disorders--beyond a simple glue in the brain. Mol Cell Neurosci 49:351–363
Weber JD, Gutmann DH (2012) Deconvoluting mTOR biology. Cell Cycle 11:236–248
Winham SJ, Cuellar-Barboza AB, McElroy SL et al (2014) Bipolar disorder with comorbid binge eating history: a genome-wide association study implicates APOB. J Affect Disord 165:151–158
Qin M, Kang J, Smith CB (2002) Increased rates of cerebral glucose metabolism in a mouse model of fragile X mental retardation. Proc Natl Acad Sci U S A 99:15758–15763
Besshoh S, Bawa D, Teves L et al (2005) Increased phosphorylation and redistribution of NMDA receptors between synaptic lipid rafts and post-synaptic densities following transient global ischemia in the rat brain. J Neurochem 93:186–194
Acknowledgments
We thank to the patient’s parents for their continuous cooperation. We thank to Dr. V. Kalscheuer and Dr. U. Reinhard for their important work to help us to refine the translocation breakpoints and set up the LCLs. This work was supported by PROMEP (No. 103.5/11/4330), PAICYT (No. CS-927-11), and CONACYT (No. INFRA-2013-204423) for C Córdova-Fletes. I. Delint-Ramírez was supported by CONACYT (No. 180919). We also thank Dr. H. Rivera for his support to review this manuscript, and B. Verduzco-Garza, E.N. Garza-Treviño, and A. Camacho for their technical support/suggestions.
Conflict of interest
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(XLS 119 kb)
Rights and permissions
About this article
Cite this article
Córdova-Fletes, C., Domínguez, M.G., Delint-Ramirez, I. et al. A de novo t(10;19)(q22.3;q13.33) leads to ZMIZ1/PRR12 reciprocal fusion transcripts in a girl with intellectual disability and neuropsychiatric alterations. Neurogenetics 16, 287–298 (2015). https://doi.org/10.1007/s10048-015-0452-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-015-0452-2