Abstract
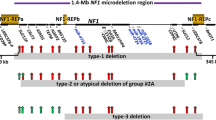
Neurofibromatosis type 1 (NF1) is a common genetic disease caused by haploinsufficiency of the NF1 tumor-suppressor gene. Different pathogenetic mechanisms have been identified, with the majority (95%) causing intragenic lesions. Single or multiexon NF1 copy number changes occur in about 2% of patients, but little is known about the molecular mechanisms behind these intragenic deletions. We report here on the molecular characterization of a novel NF1 multiexonic deletion. The application of a multidisciplinary approach including multiplex ligation-dependent probe amplification, allelic segregation analysis, and fluorescent in situ hybridization allowed us to map the breakpoints in IVS27b and IVS48. Furthermore, the breakpoint junction was characterized by sequencing. Using bioinformatic analysis, we identified some recombinogenic motifs in close proximity to the centromeric and telomeric breakpoints and predicted the presence of a mutated messenger ribonucleic acid, which was deleted between exons 28 and 48 and encodes a neurofibromin that lacks some domains essential for its function. Through reverse transcriptase–polymerase chain reaction, the expression of the mutated allele was verified, showing the junction between exons 27b and 49 and, as expected, was not subjected to nonsense-mediated decay. Multiexonic deletions represent 2% of NF1 mutations, and until now, the breakpoint has been identified in only a few cases. The fine characterization of multiexonic deletions broadens the mutational repertoire of the NF1 gene, allowing for the identification of different pathogenetic mechanisms causing NF1.


Similar content being viewed by others
References
Cawthon RM, O’Connell P, Buchberg AM, Viskochil D, Weiss RB, Culver M, Stevens J, Jenkins NA, Copeland NG, White R (1990) Identification and characterization of transcripts from the neurofibromatosis 1 region: the sequence and genomic structure of EVI2 and mapping of other transcripts. Genomics 7:555–565
Wallace M, Marchuk D, Andersen L, Letcher R, Odeh H, Saulino A, Fountain J, Brereton A, Nicholson J, Mitchell AL (1990) Type 1 neurofibromatosis gene: identification of a large transcript disrupted in three NF1 patients. Science 249:181–186
Riva P, Corrado L, Natacci F, Castorina P, Wu BL, Schneider GH, Clementi M, Tenconi R, Korf BR, Larizza L (2000) NF1 microdeletion syndrome: refined FISH characterization of sporadic and familial deletions with locus-specific probes. Am J Hum Genet 66:100–109
Jenne DE, Tinschert S, Reimann H, Lasinger W, Thiel G, Hameister H, Kehrer-Sawatzki H (2001) Molecular characterization and gene content of breakpoint boundaries in patients with neurofibromatosis type 1 with 17q11.2 microdeletions. Am J Hum Genet 69:516–527
Fang LJ, Vidaud D, Vidaud M, Thirion JP (2001) Identification and characterization of four novel large deletions in the human neurofibromatosis type 1 (NF1) gene. Hum Mutat 18:549–550
Wimmer K, Yao S, Claes K, Kehrer-Sawatzki H, Tinschert S, De Raedt T, Legius E, Callens T, Beiglbock H, Maertens O, Messiaen L (2006) Spectrum of single- and multiexon NF1 copy number changes in a cohort of 1,100 unselected NF1 patients. Genes Chromosomes Cancer 45:265–276
Dorschner MO, Sybert VP, Weaver M, Pletcher BA, Stephens K (2000) NF1 microdeletion breakpoints are clustered at flanking repetitive sequences. Hum Mol Genet 9:35–46
Liu MT, Su JS, Huang CY, Tsai SF (2003) Novel mutations involving the NF1 gene coding sequence in neurofibromatosis type 1 patients from Taiwan. J Hum Genet 48:545–549
Fang L, Chalhoub N, Li W, Feingold J, Ortenberg J, Lemieux B, Thirion JP (2001) Genotype analysis of the NF1 gene in the French Canadians from the Quebec population. Am J Med Genet 104:189–198
Xu GF, Nelson L, O'Connell P, White R (1991) An Alu polymorphism intragenic to the neurofibromatosis type 1 gene (NF1). Nucleic Acids Res 19:3764
Lazaro C, Gaona A, Estivill X (1994) Two CA/GT repeat polymorphisms in intron 27 of the human neurofibromatosis (NF1) gene. Hum Genet 93:351–352
Valero MC, Velasco E, Valero A, Moreno F, Hernández-Chico C (1996) Linkage disequilibrium between four intragenic polymorphic microsatellites of the NF1 gene and its implications for genetic counselling. J Med Genet 33:590–593
Lazaro C, Gaona A, Xu G, Weiss R, Estivill X (1993) A highly informative CA/GT repeat polymorphism in intron 38 of the human neurofibromatosis type 1 (NF1) gene. Hum Genet 92:429–430
Lichter P, Cremer T (1992) Chromosome analysis by non-isotopic in situ hybridization.. In: Rooney DE, Czepulkowski BH (eds) Human cytogenetics: a practical approach: constitutional analysis. vol. 1. IRL, Oxford
Gervasini C, Venturin M, Orzan F, Friso A, Clementi M, Tenconi R, Larizza L, Riva P (2005) Uncommon Alu-mediated NF1 microdeletion with a breakpoint inside the NF1 gene. Genomics 85:273–279
Abeysinghe SS, Chuzhanova N, Krawczak M, Ball EV, Cooper DN (2003) Translocation and gross deletion breakpoints in human inherited disease and cancer I: nucleotide composition and recombination-associated motifs. Hum Mutat 22:229–244
D'Angelo I, Welti S, Bonneau F, Scheffzek K (2006) A novel bipartite phospholipid-binding module in the neurofibromatosis type 1 protein. EMBO Rep 7:174–179
Hiatt KK, Ingram DA, Zhang Y, Bollag G, Clapp DW (2001) Neurofibromin GTPase-activating protein-related domains restore normal growth in Nf1−/− cells. J Biol Chem 276:7240–7245
Thomas SL, Deadwyler GD, Tang J, Stubbs EB Jr, Muir D, Hiatt KK, Clapp DW, De Vries GH (2006) Reconstitution of the NF1 GAP-related domain in NF1-deficient human Schwann cells. Biochem Biophys Res Commun 348:971–980
Welti S, Fraterman S, D’Angelo I, Wilm M, Scheffzek K (2007) The sec14 homology module of neurofibromin binds cellular glycerophospholipids: mass spectrometry and structure of a lipid complex. J Mol Biol 366:551–562
Hsueh YP, Roberts AM, Volta M, Sheng M, Roberts RG (2001) Bipartite interaction between neurofibromatosis type I protein (neurofibromin) and syndecan transmembrane heparan sulfate proteoglycans. Neuroscience 21:3764–3770
Lin YL, Lei YT, Hong CJ, Hsueh YP (2007) Syndecan-2 induces filopodia and dendritic spine formation via the neurofibromin-PKA-E na/VASP pathway. J Cell Biol 177:829–841
Acknowledgments
This work was supported by FIRST, Italy, to PR and by the Spanish Lay Group of Neurofibromatosis (AANF).
Author information
Authors and Affiliations
Corresponding author
Additional information
Francesca Orzan and Michela Stroppi contributed equally to this work.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Orzan, F., Stroppi, M., Venturin, M. et al. Breakpoint characterization of a novel NF1 multiexonic deletion: a case showing expression of the mutated allele. Neurogenetics 9, 95–100 (2008). https://doi.org/10.1007/s10048-007-0115-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-007-0115-z