Abstract
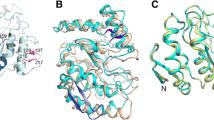
The UV-light damage specific DNA glycosylase from Chlorella virus strain PBCV-1 (pyrimidine dimer glycosylase; PDG) incises DNA at sites containing UV-induced thymidine dimers by catalyzing the breakage of the N-C-1′ glycosyl bond. As the amino acid sequence of PDG is 41% identical to that of T4 endonuclease V (Endo V), and potential key active site residues are conserved, we used coordinates from a crystal structure of an Endo V complexed with DNA containing a cis-syn cyclobutane thymidine-dimer as a template to model a similar complex of PDG. Quantum mechanical calculations of the damaged base pair and the distance geometry based program DIAMOD were used to generate a PDG/DNA model whose backbone root mean square deviation (RMSD) to the Endo V/DNA structure was 0.5 Å, 0.5 Å, and 0.8 Å for DNA, protein, and the whole complex, respectively. To better understand structural details that could account for differences in activity of the two enzymes, molecular dynamics simulations were used to follow protein-DNA interactions in an aqueous environment. The simulations of the Endo V/DNA complex indicate new roles for Arg22 and Arg26 in the active site in recognizing irregular pairing and maintaining the strand separation needed for incision of the damaged bases. The model for the PDG/DNA complex and simulations thereof indicate a similar mechanism for DNA binding by this enzyme despite significant differences in residues maintaining the flipped-out adenine and strand separation in the area of damage. According to our model, PDG′s increased affinity for substrate is probably due to a higher surface charge. Further, reduced packing density in the active site could account for PDG′s activity on trans-syn II cyclobutane dimers.
Similar content being viewed by others
Author information
Authors and Affiliations
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Zhu, H., Schein, C. & Braun, W. Homology Modeling and Molecular Dynamics Simulations of PBCV-1 Glycosylase Complexed with UV-damaged DNA. J Mol Model 5, 302–316 (1999). https://doi.org/10.1007/s0089490050302
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s0089490050302