Abstract
Context
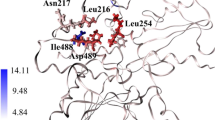
SHP2 is a non-receptor protein tyrosine phosphatase to remove tyrosine phosphorylation. Functionally, SHP2 is an essential bridge to connect numerous oncogenic cell-signaling cascades including RAS-ERK, PI3K-AKT, JAK-STAT, and PD-1/PD-L1 pathways. This study aims to discover novel and potent SHP2 inhibitors using a hierarchical structure-based virtual screening strategy that combines molecular docking and the fragment molecular orbital method (FMO) for calculating binding affinity (referred to as the Dock-FMO protocol). For the SHP2 target, the FMO method prediction has a high correlation between the binding affinity of the protein–ligand interaction and experimental values (R2 = 0.55), demonstrating a significant advantage over the MM/PBSA (R2 = 0.02) and MM/GBSA (R2 = 0.15) methods. Therefore, we employed Dock-FMO virtual screening of ChemDiv database of ∼2,990,000 compounds to identify a novel SHP2 allosteric inhibitor bearing hydroxyimino acetamide scaffold. Experimental validation demonstrated that the new compound (E)-2-(hydroxyimino)-2-phenyl-N-(piperidin-4-ylmethyl)acetamide (7188–0011) effectively inhibited SHP2 in a dose-dependent manner. Molecular dynamics (MD) simulation analysis revealed the binding stability of compound 7188–0011 and the SHP2 protein, along with the key interacting residues in the allosteric binding site. Overall, our work has identified a novel and promising allosteric inhibitor that targets SHP2, providing a new starting point for further optimization to develop more potent inhibitors.
Methods
All the molecular docking studies were employed to identify potential leads with Maestro v10.1. The protein–ligand binding affinities of potential leads were further predicted by FMO calculations at MP2/6-31G* level using GAMESS v2020 system. MD simulations were carried out with AmberTools18 by applying the FF14SB force field. MD trajectories were analyzed using VMD v1.9.3. MM/GB(PB)SA binding free energy analysis was carried out with the mmpbsa.py tool of AmberTools18. The docking and MD simulation results were visualized through PyMOL v2.5.0.




Similar content being viewed by others
Data Availability
FMO calculation was applied using FMO code version 5.1 as embedded in General Atomic and Molecular Electronic Structure System (GAMESS), available at https://www.msg.chem.iastate.edu/gamess/[M1] [z2] (accessed on 21 May 2022). Calculated energy values are summarized in the Supporting Information and Supplemental Data.
References
Chan RJ, Feng GS (2007) PTPN11 is the first identified proto-oncogene that encodes a tyrosine phosphatase. Blood 109(3):862–867
Grossmann KS, Rosário M, Birchmeier C, Birchmeier W (2010) Chapter 2 - The tyrosine phosphatase Shp2 in development and cancer. In Adv. Cancer Res., Vande Woude, G. F.; Klein, G., Eds. Academic Press: Vol. 106, pp 53–89
Barr AJ, Ugochukwu E, Lee WH, King ON, Filippakopoulos P, Alfano I, Savitsky P, Burgess-Brown NA, Müller S, Knapp S (2009) Large-scale structural analysis of the classical human protein tyrosine phosphatome. Cell 136(2):352–363
Neel BG, Gu H, Pao L (2003) The ’Shp’ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem Sci 28(6):284–293
Tiganis T, Bennett AM (2007) Protein tyrosine phosphatase function: the substrate perspective. Biochem J 402(1):1–15
Vazhappilly CG, Saleh E, Ramadan W, Menon V, Al-Azawi AM, Tarazi H, Abdu-Allah H, El-Shorbagi AN, El-Awady R (2019) Inhibition of SHP2 by new compounds induces differential effects on RAS/RAF/ERK and PI3K/AKT pathways in different cancer cell types. Invest New Drugs 37(2):252–261
Wong GS, Zhou J, Liu JB, Wu Z, Xu X, Li T, Xu D, Schumacher SE, Puschhof J, McFarland J, Zou C, Dulak A, Henderson L, Xu P, O’Day E, Rendak R, Liao WL, Cecchi F, Hembrough T, Schwartz S, Szeto C, Rustgi AK, Wong KK, Diehl JA, Jensen K, Graziano F, Ruzzo A, Fereshetian S, Mertins P, Carr SA, Beroukhim R, Nakamura K, Oki E, Watanabe M, Baba H, Imamura Y, Catenacci D, Bass AJ (2018) Targeting wild-type KRAS-amplified gastroesophageal cancer through combined MEK and SHP2 inhibition. Nat Med 24(7):968–977
Gavrieli M, Watanabe N, Loftin SK, Murphy TL, Murphy KM (2003) Characterization of phosphotyrosine binding motifs in the cytoplasmic domain of B and T lymphocyte attenuator required for association with protein tyrosine phosphatases SHP-1 and SHP-2. Biochem Biophys Res Commun 312(4):1236–1243
Yokosuka T, Takamatsu M, Kobayashi-Imanishi W, Hashimoto-Tane A, Azuma M, Saito T (2012) Programmed cell death 1 forms negative costimulatory microclusters that directly inhibit T cell receptor signaling by recruiting phosphatase SHP2. J Exp Med 209(6):1201–1217
Chemnitz JM, Parry RV, Nichols KE, June CH, Riley JL (2004) SHP-1 and SHP-2 associate with immunoreceptor tyrosine-based switch motif of programmed death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation. J Immunol (Baltimore Md 1950) 173(2):945–54
Song Y, Zhao M, Zhang H, Yu B (2022) Double-edged roles of protein tyrosine phosphatase SHP2 in cancer and its inhibitors in clinical trials. Pharmacol Ther 230:107966
Miyamoto D, Miyamoto M, Takahashi A, Yomogita Y, Higashi H, Kondo S, Hatakeyama M (2008) Isolation of a distinct class of gain-of-function SHP-2 mutants with oncogenic RAS-like transforming activity from solid tumors. Oncogene 27(25):3508–3515
Chan G, Kalaitzidis D, Neel BG (2008) The tyrosine phosphatase Shp2 (PTPN11) in cancer. Cancer Metastasis Rev 27(2):179–192
Grosskopf S, Eckert C, Arkona C, Radetzki S, Böhm K, Heinemann U, Wolber G, von Kries JP, Birchmeier W, Rademann J (2015) Selective inhibitors of the protein tyrosine phosphatase SHP2 block cellular motility and growth of cancer cells in vitro and in vivo. ChemMedChem 10(5):815–826
Chen C, Liang F, Chen B, Sun Z, Xue T, Yang R, Luo D (2017) Identification of demethylincisterol A(3) as a selective inhibitor of protein tyrosine phosphatase Shp2. Eur J Pharmacol 795:124–133
Chen C, Cao M, Zhu S, Wang C, Liang F, Yan L, Luo D (2015) Discovery of a novel inhibitor of the protein tyrosine phosphatase Shp2. Sci Rep 5:17626
Chen L, Sung SS, Yip ML, Lawrence HR, Ren Y, Guida WC, Sebti SM, Lawrence NJ, Wu J (2006) Discovery of a novel shp2 protein tyrosine phosphatase inhibitor. Mol Pharmacol 70(2):562–570
Nören-Müller A, Reis-Corrêa I, Prinz H Jr, Rosenbaum C, Saxena K, Schwalbe HJ, Vestweber D, Cagna G, Schunk S, Schwarz O, Schiewe H, Waldmann H (2006) Discovery of protein phosphatase inhibitor classes by biology-oriented synthesis. Proc Natl Acad Sci U S A 103(28):10606–11
Chen YN, LaMarche MJ, Chan HM, Fekkes P, Garcia-Fortanet J, Acker MG, Antonakos B, Chen CH, Chen Z, Cooke VG, Dobson JR, Deng Z, Fei F, Firestone B, Fodor M, Fridrich C, Gao H, Grunenfelder D, Hao HX, Jacob J, Ho S, Hsiao K, Kang ZB, Karki R, Kato M, Larrow J, La Bonte LR, Lenoir F, Liu G, Liu S, Majumdar D, Meyer MJ, Palermo M, Perez L, Pu M, Price E, Quinn C, Shakya S, Shultz MD, Slisz J, Venkatesan K, Wang P, Warmuth M, Williams S, Yang G, Yuan J, Zhang JH, Zhu P, Ramsey T, Keen NJ, Sellers WR, Stams T, Fortin PD (2016) Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature 535(7610):148–152
Bhardwaj V, Purohit R (2020) Computational investigation on effect of mutations in PCNA resulting in structural perturbations and inhibition of mismatch repair pathway. J Biomol Struct Dyn 38(7):1963–1974
Rajasekaran R, George Priya Doss C, Sudandiradoss C, Ramanathan K, Purohit R, Sethumadhavan R (2008) Effect of deleterious nsSNP on the HER2 receptor based on stability and binding affinity with herceptin: a computational approach. CR Biol 331(6):409–17
Singh R, Bhardwaj V, Purohit R (2021) Identification of a novel binding mechanism of Quinoline based molecules with lactate dehydrogenase of Plasmodium falciparum. J Biomol Struct Dyn 39(1):348–356
Kamaraj B, Purohit R (2016) Mutational analysis on membrane associated transporter protein (MATP) and their structural consequences in oculocutaeous albinism type 4 (OCA4)-a molecular dynamics approach. J Cell Biochem 117(11):2608–2619
Bhardwaj VK, Purohit R (2020) A new insight into protein-protein interactions and the effect of conformational alterations in PCNA. Int J Biol Macromol 148:999–1009
Kumar S, Sinha K, Sharma R, Purohit R, Padwad Y (2019) Phloretin and phloridzin improve insulin sensitivity and enhance glucose uptake by subverting PPARγ/Cdk5 interaction in differentiated adipocytes. Exp Cell Res 383(1):111480
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS (2004) Glide: A new approach for rapid, accurate docking and scoring. 1. Method and Assessment of Docking Accuracy. J Med Chem 47(7):1739–1749
Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL (2004) Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment Factors in Database Screening. J Med Chem 47(7):1750–1759
Fedorov DG, Kitaura K (2007) Extending the power of quantum chemistry to large systems with the fragment molecular orbital method. J Phys Chem A 111(30):6904–6914
Fedorov DG, Nagata T, Kitaura K (2012) Exploring chemistry with the fragment molecular orbital method. Phys Chem Chem Phys 14(21):7562–7577
Fedorov DG (2017) The fragment molecular orbital method: theoretical development, implementation in GAMESS, and applications. WIREs Comput Mol Sci 7(6):e1322
Schmidt MW, Baldridge KK, Boatz JA, Elbert ST, Gordon MS, Jensen JH, Koseki S, Matsunaga N, Nguyen KA, Su S, Windus TL, Dupuis M, Montgomery JA (1993) General atomic and molecular electronic structure system. J Comput Chem 14(11):1347–1363
Fedorov DG, Kitaura K (2007) Pair interaction energy decomposition analysis. J Comput Chem 28(1):222–237
LaMarche MJ, Acker M, Argintaru A, Bauer D, Boisclair J, Chan H, Chen CH, Chen YN, Chen Z, Deng Z, Dore M, Dunstan D, Fan J, Fekkes P, Firestone B, Fodor M, Garcia-Fortanet J, Fortin PD, Fridrich C, Giraldes J, Glick M, Grunenfelder D, Hao HX, Hentemann M, Ho S, Jouk A, Kang ZB, Karki R, Kato M, Keen N, Koenig R, LaBonte LR, Larrow J, Liu G, Liu S, Majumdar D, Mathieu S, Meyer MJ, Mohseni M, Ntaganda R, Palermo M, Perez L, Pu M, Ramsey T, Reilly J, Sarver P, Sellers WR, Sendzik M, Shultz MD, Slisz J, Slocum K, Smith T, Spence S, Stams T, Straub C, Tamez V, Toure BB Jr, Towler C, Wang P, Wang H, Williams SL, Yang F, Yu B, Zhang JH, Zhu S (2020) Identification of TNO155, an allosteric SHP2 inhibitor for the treatment of cancer. J Med Chem 63(22):13578–13594
Sriwilaijaroen N, Magesh S, Imamura A, Ando H, Ishida H, Sakai M, Ishitsubo E, Hori T, Moriya S, Ishikawa T, Kuwata K, Odagiri T, Tashiro M, Hiramatsu H, Tsukamoto K, Miyagi T, Tokiwa H, Kiso M, Suzuki Y (2016) A novel potent and highly specific inhibitor against influenza viral N1–N9 neuraminidases: insight into neuraminidase-inhibitor interactions. J Med Chem 59(10):4563–4577
Li S, Qin C, Cui S, Xu H, Wu F, Wang J, Su M, Fang X, Li D, Jiao Q, Zhang M, Xia C, Zhu L, Wang R, Li J, Jiang H, Zhao Z, Li J, Li H (2019) Discovery of a natural-product-derived preclinical candidate for once-weekly treatment of type 2 diabetes. J Med Chem 62(5):2348–2361
Heifetz A, Chudyk EI, Gleave L, Aldeghi M, Cherezov V, Fedorov DG, Biggin PC, Bodkin MJ (2016) The fragment molecular orbital method reveals new insight into the chemical nature of GPCR-ligand interactions. J Chem Inf Model 56(1):159–172
Heifetz A, Trani G, Aldeghi M, MacKinnon CH, McEwan PA, Brookfield FA, Chudyk EI, Bodkin M, Pei Z, Burch JD, Ortwine DF (2016) Fragment molecular orbital method applied to lead optimization of novel interleukin-2 inducible T-cell kinase (ITK) inhibitors. J Med Chem 59(9):4352–4363
Mazanetz MP, Ichihara O, Law RJ, Whittaker M (2011) Prediction of cyclin-dependent kinase 2 inhibitor potency using the fragment molecular orbital method. J Cheminform 3(1):2
Fedorov DG, Kitaura K (2007) Pair interaction energy decomposition analysis. J Comput Chem 28(1):222–237
Sarver P, Acker M, Bagdanoff JT, Chen Z, Chen YN, Chan H, Firestone B, Fodor M, Fortanet J, Hao H, Hentemann M, Kato M, Koenig R, LaBonte LR, Liu G, Liu S, Liu C, McNeill E, Mohseni M, Sendzik M, Stams T, Spence S, Tamez V, Tichkule R, Towler C, Wang H, Wang P, Williams SL, Yu B, LaMarche MJ (2019) 6-Amino-3-methylpyrimidinones as potent, selective, and orally efficacious SHP2 inhibitors. J Med Chem 62(4):1793–1802
Bagdanoff JT, Chen Z, Acker M, Chen Y-N, Chan H, Dore M, Firestone B, Fodor M, Fortanet J, Hentemann M, Kato M, Koenig R, La Bonte LR, Liu S, Mohseni M, Ntaganda R, Sarver P, Smith T, Sendzik M, Stams T, Spence S, Towler C, Wang H, Wang P, Williams SL, La Marche MJ (2019) Optimization of fused bicyclic allosteric SHP2 inhibitors. J Med Chem 62(4):1781–1792
LaRochelle JR, Fodor M, Ellegast JM, Liu X, Vemulapalli V, Mohseni M, Stams T, Buhrlage SJ, Stegmaier K, LaMarche MJ, Acker MG, Blacklow SC (2017) Identification of an allosteric benzothiazolopyrimidone inhibitor of the oncogenic protein tyrosine phosphatase SHP2. Bioorg Med Chem 25(24):6479–6485
Garcia Fortanet J, Chen CH-T, Chen Y-NP, Chen Z, Deng Z, Firestone B, Fekkes P, Fodor M, Fortin PD, Fridrich C, Grunenfelder D, Ho S, Kang ZB, Karki R, Kato M, Keen N, LaBonte LR, Larrow J, Lenoir F, Liu G, Liu S, Lombardo F, Majumdar D, Meyer MJ, Palermo M, Perez L, Pu M, Ramsey T, Sellers WR, Shultz MD, Stams T, Towler C, Wang P, Williams SL, Zhang J-H, LaMarche MJ (2016) Allosteric inhibition of SHP2: identification of a potent, selective, and orally efficacious phosphatase inhibitor. J Med Chem 59(17):7773–7782
Funding
This work was supported in part by the National Key R&D Program of China (2022YFC3400504), the National Natural Science Foundation of China (grants 82173690 to S.L.L. and 81825020 to H.L.,), and the Fundamental Research Funds for the Central Universities; S.L.L. is also sponsored by the Shanghai Rising-Star Program (23QA1402800). Honglin Li was also sponsored by the National Program for Special Supports of Eminent Professionals and National Program for Support of Top-Notch Young Professionals.
Author information
Authors and Affiliations
Contributions
Z.Y., M.Z., and L.C. performed research and drafted the manuscript; they contributed equally to the study. Z.Y. and M.Z. wrote the main manuscript, and L.C. conducted protein expression, purification, and biochemical assay. X.C., S.R., S.S., and Y.Z. performed the data collection. S.L., H.L., and L.Z. conceived and supervised the overall project. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yuan, Z., Zhang, M., Chang, L. et al. Discovery of a novel SHP2 allosteric inhibitor using virtual screening, FMO calculation, and molecular dynamic simulation. J Mol Model 30, 131 (2024). https://doi.org/10.1007/s00894-024-05935-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-024-05935-y