Abstract
Context
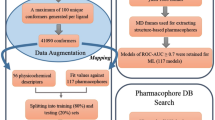
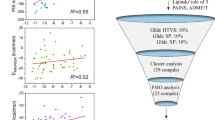
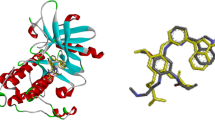
The Hedgehog (Hh) signaling pathway is a crucial regulator of various cellular processes. Dysregulated activation of the Smoothened (SMO) oncoprotein, a key component of the Hh pathway, has been implicated in several types of cancer. Although SMO inhibitors are important anti-cancer therapeutics, drug-resistant SMO mutants have emerged, limiting their efficacy. This study aimed to discover stable SMO inhibitors for both wild-type and mutant SMOs, using a 12-feature pharmacophore model validated for virtual screening. One lead compound, LCT10312, was identified with high affinity to SMO and showed a significant conformational change in the SMO structure upon binding. Molecular dynamic simulation revealed stable interaction of LCT10312 with SMO and large atom motions, indicating SMO structural fluctuation. The lead compound showed high predicted binding scores to several clinically relevant SMO mutants.
Methods
A ligand-based pharmacophore model was developed from 25 structurally clustered SMO inhibitors using LigandScout v3.12 software and virtually screened for hit identification from a library of 511,878 chemicals. Molecular docking was employed to identify potential leads based on SMO affinities. Molecular dynamic simulation (MDS) with GROMACS v5.1.4 was performed to analyze the structural changes of SMO oncoprotein upon binding lead compound(s) and cyclopamine as the control for 100 ns. The binding affinity of lead compound(s) was predicted on clinical and laboratory SMO mutants.






Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Finco I, LaPensee CR, Krill KT, Hammer GD (2015) Hedgehog signaling and steroidogenesis. Annu Rev Physiol 77:105–129. https://doi.org/10.1146/annurev-physiol-061214-111754
Jeng KS, Sheen IS, Leu CM et al (2020) The role of smoothened in cancer. Int J Mol Sci 21:1–20. https://doi.org/10.3390/ijms21186863
Riobo NA, Manning DR (2007) Pathways of signal transduction employed by vertebrate Hedgehogs. Biochem J 403:369–379. https://doi.org/10.1042/BJ20061723
Skoda AM, Simovic D, Karin V et al (2018) The role of the hedgehog signaling pathway in cancer: a comprehensive review. Bosn J Basic Med Sci 18:8–20. https://doi.org/10.17305/bjbms.2018.2756
Carballo GB, Honorato JR, De Lopes GPF, Spohr TCLDSE (2018) A highlight on Sonic hedgehog pathway. Cell Commun Signal 16:1–15. https://doi.org/10.1186/s12964-018-0220-7
Jamieson C, Martinelli G, Papayannidis C, Cortes JE (2020) Hedgehog pathway inhibitors: a new therapeutic class for the treatment of acute myeloid leukemia. Blood Cancer Discov 1:134–145. https://doi.org/10.1158/2643-3230.bcd-20-0007
Niewiadomski P, Niedziółka SM, Markiewicz Ł et al (2019) Gli proteins: regulation in development and cancer. Cells 8:147. https://doi.org/10.3390/cells8020147
Iriana S, Asha K, Repak M, Sharma-Walia N (2021) Hedgehog signaling: implications in cancers and viral infections. Int J Mol Sci 22:1–30. https://doi.org/10.3390/ijms22031042
Zhang H, Sun Z, Liu Z, Song C (2018) Overcoming the emerging drug resistance of smoothened: an overview of small-molecule SMO antagonists with antiresistance activity. Future Med Chem 10:2855–2875. https://doi.org/10.4155/fmc-2018-0200
Sun C, Zhang Y, Wang H et al (2021) Design and biological evaluation of phenyl imidazole analogs as hedgehog signaling pathway inhibitors. Chem Biol Drug Des 97:546–552. https://doi.org/10.1111/cbdd.13799
Quaglio D, Infante P, Di Marcotullio L et al (2020) Hedgehog signaling pathway inhibitors: an updated patent review (2015–present). Expert Opin Ther Pat 30:235–250. https://doi.org/10.1080/13543776.2020.1730327
Wei SF, He DH, Zhang SB et al (2021) Identification of pseudolaric acid B as a novel Hedgehog pathway inhibitor in medulloblastoma. Biochem Pharmacol 190:114593. https://doi.org/10.1016/j.bcp.2021.114593
Fan J, Li H, Kuang L et al (2021) Identification of a potent antagonist of smoothened in hedgehog signaling. Cell Biosci 11:1–11. https://doi.org/10.1186/s13578-021-00558-9
Weierstall U, James D, Wang C et al (2014) Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nat Commun 5:1–6. https://doi.org/10.1038/ncomms4309
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723. https://doi.org/10.1002/elps.1150181505
Mohebbi A, Mohammadi S, Memarian A (2016) Prediction of HBF-0259 interactions with hepatitis B virus receptors and surface antigen secretory factors. VirusDisease 27:234–241. https://doi.org/10.1007/s13337-016-0333-9
Pettersen EF, Goddard TD, Huang CC et al (2004) UCSF Chimera - a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. https://doi.org/10.1002/jcc.20084
Mohebbi A, Mirarab A, Shaddel R et al (2021) Molecular dynamic simulation and docking of cyclophilin A mutants with its potential inhibitors. J Clin Basic Res 5:26–41. https://doi.org/10.52547/jcbr.5.2.26
Dunbrack RL (2002) Rotamer libraries in the 21st century. Curr Opin Struct Biol 12:431–440. https://doi.org/10.1016/S0959-440X(02)00344-5
Askari FS, Ebrahimi M, Parhiz J et al (2022) Digging for the discovery of SARS-CoV-2 nsp12 inhibitors: a pharmacophore-based and molecular dynamics simulation study. Future Virol. https://doi.org/10.2217/fvl-2022-0054
Wolber G, Langer T (2005) LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J Chem Inf Model 45:160–169. https://doi.org/10.1021/ci049885e
Pence HE, Williams A (2010) ChemSpider: an online chemical information resource. J Chem Educ 87:1123–1124. https://doi.org/10.1021/ed100697w
Bolton EE, Wang Y, Thiessen PA, Bryant SH (2008) Chapter 12 PubChem: integrated platform of small molecules and biological activities. Annu Rep Comput Chem 4:217–241. https://doi.org/10.1016/S1574-1400(08)00012-1
O’Boyle NM, Banck M, James CA et al (2011) Open Babel: an open chemical toolbox. J Cheminform 3. https://doi.org/10.1186/1758-2946-3-33
Mohebbi A, Askari FS, Sammak AS et al (2021) Druggability of cavity pockets within SARS-CoV-2 spike glycoprotein and pharmacophore-based drug discovery. Future Virol 16:389–397. https://doi.org/10.2217/fvl-2020-0394
Allouche A (2012) Software news and updates Gabedit — a graphical user interface for computational chemistry softwares. J Comput Chem 32:174–182. https://doi.org/10.1002/jcc
Abraham MJ, Murtola T, Schulz R et al (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19–25. https://doi.org/10.1016/j.softx.2015.06.001
Oostenbrink C, Villa A, Mark AE, Van Gunsteren WF (2004) A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J Comput Chem 25:1656–1676. https://doi.org/10.1002/jcc.20090
Schüttelkopf AW, Van Aalten DMF (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr Sect D Biol Crystallogr 60:1355–1363. https://doi.org/10.1107/S0907444904011679
Eisenberg D, Mclachlan AD (1986) Solvation energy in protein folding and binding. Nature 319:199–203. https://doi.org/10.1038/319199a0
Bondi A (1964) Van der Waals volumes and radii. J Phys Chem 68:441–451. https://doi.org/10.1021/j100785a001
Nasution MAF, Parikesit AA, Tambunan USF (2020) Discovery of novel drug candidates based on herbaric acid derivates as potential inhibitors of the hedgehog signaling pathway in cervical cancer therapeutics. J Phys Conf Ser 1442. https://doi.org/10.1088/1742-6596/1442/1/012052
Ekawati MM, Fardiansyah Nasution MA, Siregar S et al (2019) Pharmacophore-based virtual screening and molecular docking simulation of terpenoid compounds as the inhibitor of sonic hedgehog protein for colorectal cancer therapy. IOP Conf Ser Mater Sci Eng 509. https://doi.org/10.1088/1757-899X/509/1/012075
Manetti F, Stecca B, Santini R et al (2020) Pharmacophore-based virtual screening for identification of negative modulators of GLI1 as potential anticancer agents. ACS Med Chem Lett 11:832–838. https://doi.org/10.1021/acsmedchemlett.9b00639
Hwang S, Thangapandian S, Lee KW (2013) Molecular dynamics simulations of sonic hedgehog-receptor and inhibitor complexes and their applications for potential anticancer agent discovery. PLoS One 8. https://doi.org/10.1371/journal.pone.0068271
Lu W, Zhang D, Ma H et al (2018) Discovery of potent and novel smoothened antagonists via structure-based virtual screening and biological assays. Eur J Med Chem 155:34–48. https://doi.org/10.1016/j.ejmech.2018.05.035
Nasution MAF, Stephanie F, Tambunan USF (2019) Pharmacophore-based virtual screening and molecular docking simulation of flavonoids as smoothened protein inhibitor of Hedgehog signaling pathways. AIP Conf Proc 2168. https://doi.org/10.1063/1.5132487
Sinha N, Chowdhury S, Sarkar RR (2018) Deciphering structural stability and binding mechanisms of potential antagonists with smoothened protein. J Biomol Struct Dyn 36:2917–2937. https://doi.org/10.1080/07391102.2017.1372310
Xie J, Murone M, Luoh SM et al (1998) Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature 391:90–92. https://doi.org/10.1038/34201
Sweeney RT, McClary AC, Myers BR et al (2014) Identification of recurrent SMO and BRAF mutations in ameloblastomas. Nat Genet 46:722–725. https://doi.org/10.1038/ng.2986
Twigg SRF, Hufnagel RB, Miller KA et al (2016) A recurrent mosaic mutation in SMO, encoding the hedgehog signal transducer smoothened, is the major cause of Curry-Jones syndrome. Am J Hum Genet 98:1256–1265. https://doi.org/10.1016/j.ajhg.2016.04.007
Katoh M (2019) Genomic testing, tumor microenvironment and targeted therapy of Hedgehog-related human cancers. Clin Sci 133:953–970. https://doi.org/10.1042/CS20180845
Sharpe HJ, Pau G, Dijkgraaf GJ et al (2015) Genomic analysis of smoothened inhibitor resistance in basal cell carcinoma. Cancer Cell 27:327–341. https://doi.org/10.1016/j.ccell.2015.02.001
Bendell J, Andre V, Ho A et al (2018) Phase I study of ly2940680, a SMO antagonist, in patients with advanced cancer including treatment-naïve and previously treated basal cell carcinoma. Clin Cancer Res 24:2082–2091. https://doi.org/10.1158/1078-0432.CCR-17-0723
Atwood SX, Sarin KY, Whitson RJ et al (2015) Smoothened variants explain the majority of drug resistance in basal cell carcinoma. Cancer Cell 27:342–353. https://doi.org/10.1016/j.ccell.2015.02.002
Dijkgraaf GJP, Alicke B, Weinmann L et al (2011) Small molecule inhibition of GDC-0449 refractory smoothened mutants and downstream mechanisms of drug resistance. Cancer Res 71:435–444. https://doi.org/10.1158/0008-5472.CAN-10-2876/649585/AM/SMALL-MOLECULE-INHIBITION-OF-GDC-0449-REFRACTORY
Rudin CM, Hann CL, Laterra J et al (2009) Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med 361:1173–1178. https://doi.org/10.1056/NEJMOA0902903/SUPPL_FILE/NEJM_RUDIN_1173SA1.PDF
Li Y, Song Q, Day BW (2019) Phase I and phase II sonidegib and vismodegib clinical trials for the treatment of paediatric and adult MB patients: a systemic review and meta-analysis. Acta Neuropathol Commun 7:123. https://doi.org/10.1186/S40478-019-0773-8/FIGURES/4
Acknowledgements
The authors wish to thank the Stem Cell Research Center and Department of Microbiology for their spiritual supports.
Funding
Financial support for this study was provided by a grant (970913172).
Author information
Authors and Affiliations
Contributions
The study conception and design, as well as data gathering, analysis, visualization, writing, and editing the drafts of the manuscript are all performed by Alireza Mohebbi.
Corresponding author
Ethics declarations
Ethics approval
An ethical approval code (IR.GOUMS.REC.1397.066) received from the Golestan University of Medical Sciences, Gorgan, Iran.
Competing interests
The author declares no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mohebbi, A. Ligand-based 3D pharmacophore modeling, virtual screening, and molecular dynamic simulation of potential smoothened inhibitors. J Mol Model 29, 143 (2023). https://doi.org/10.1007/s00894-023-05532-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-023-05532-5