Abstract
Context
Lymphatic filariasis, generally called as elephantiasis, is a vector-borne infectious disease caused by the filarial nematodes, mainly Wuchereria bancrofti, Brugia malayi, and Brugia timori, which are transmitted through mosquitoes. The infection affects the normal flow of lymph leading to abnormal enlargement of body parts, severe pain, permanent disability, and social stigma. Due to the development of resistance as well as toxic effects, existing medicines for lymphatic filariasis are becoming ineffective in killing the adult worms. It is essential to search novel filaricidal drugs with new molecular targets. Asparaginyl-tRNA synthetase (PDB ID: 2XGT) belongs to the group of aminoacyl-tRNA synthetases that catalyze specific attachment of amino acids to their tRNA during protein biosynthesis. Plants and their extracts are well-known medicinal practice for the management of several parasitic infectious diseases including filarial infections.
Methods
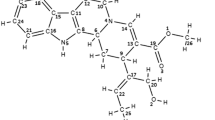
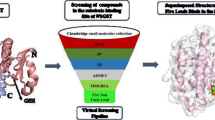
In this study, asparaginyl-tRNA synthetase of Brugia malayi was used as a target to perform virtual screening of plant phytoconstituents of Vitex negundo from IMPPAT database, which exhibits anti-filarial and anti-helminthic properties. A total of sixty-eight compounds from Vitex negundo were docked against asparaginyl-tRNA synthetase using Autodock module of PyRx tool. Among the 68 compounds screened, 3 compounds, negundoside, myricetin, and nishindaside, exhibited a higher binding affinity compared to standard drugs. The pharmacokinetic and physicochemical prediction, stability of ligand-receptor complexes via molecular dynamics simulation, and density functionality theory were done further for the top-scored ligands with receptor.









Similar content being viewed by others
Data availability
Not applicable.
Code availability
This work was conducted using the free software Autodock Vina module in PyRx 0.8 software and GROMACS.
References
Cromwell EA, Schmidt CA, Kwong KT, Pigott DM, Mupfasoni D, Biswas G, Shirude S, Hill E, Donkers KM, Abdoli A (2020) The global distribution of lymphatic filariasis, 2000–18: a geospatial analysis. Lancet Glob Health 8:e1186–e1194. https://doi.org/10.1016/S2214-109X(20)30286-2
Chandy A, Thakur AS, Singh MP, Manigauha A (2011) A review of neglected tropical diseases: filariasis. Asian Pac J Trop Med 4:581–586. https://doi.org/10.1016/S1995-7645(11)60150-8
Joshi, P.: Epidemiology of lymphatic filariasis. In Lymphatic filariasis; Springer: 1–14. (2018). https://doi.org/10.1007/978-981-13-1391-2_1
Anil N, Talluri VR (2015) Lymphatic filariasis: drug targets and nematicidal plants. J Pharm Sci Res 7:928
Yamamoto T, Daniel BW, Rodriguez JR, Kageyama T, Sakai H, Fuse Y, Tsukuura R, Yamamoto N (2022) Radical reduction and reconstruction for male genital elephantiasis: superficial circumflex iliac artery perforator (SCIP) lymphatic flap transfer after elephantiasis tissue resection. J Plast Reconstr Aesthet Surg 75:870–880. https://doi.org/10.1016/j.bjps.2021.08.011
Kamgno J, Djeunga HN (2020) Progress towards global elimination of lymphatic filariasis. Lancet Glob Health 8:e1108–e1109. https://doi.org/10.1016/S2214-109X(20)30323-5
Geary TG, Haque M (2021) Human helminth infections: a primer. In: Humphries DL, Scott ME, Vermund H (eds) Nutrition and Infectious Diseases. Springer, Cham, pp 189–215. https://doi.org/10.1007/978-3-030-56913-6_7
Schorderet-Weber S, Noack S, Selzer PM, Kaminsky R (2017) Blocking transmission of vector-borne diseases. Int J Parasitol Drugs Drug Resist 7:90–109. https://doi.org/10.1002/9783527802883.ch3
Arya H, Coumar MS (2014) Virtual screening of traditional Chinese medicine (TCM) database: identification of fragment-like lead molecules for filariasis target asparaginyl-tRNA synthetase. J Mol Model 20:1–13. https://doi.org/10.1007/s00894-014-2266-9
Alitalo K, Carmeliet P (2002) Molecular mechanisms of lymphangiogenesis in health and disease. Cancer Cell 1:219–227. https://doi.org/10.1016/S1535-6108(02)00051-X
Goa KL, McTavish D, Clissold SP (1991) Ivermectin Drugs 42:640–658. https://doi.org/10.2165/00003495-199142040-00007
Ottesen E, Ismail M, Horton J (1999) The role of albendazole in programmes to eliminate lymphatic filariasis. Parasitol Today 15:382–386. https://doi.org/10.1016/S0169-4758(99)01486-6
Misra-Bhattacharya S, Shahab M (2018) Progress in the treatment and control of lymphatic filariasis. In Lymphatic Filariasis; Epidemiology, Treatment and Prevention - The Indian Perspective, Springer, pp 47–58. https://doi.org/10.1007/978-981-13-1391-2_4
Chandrasekar R, Sivanesan S, Natarajan M, Naveena K, Preetha N, Karthika S, Vimalraj S, Kron M, Dhanasekaran A (2021) Evaluation of the angiogenic properties of Brugia malayi asparaginyl-tRNA synthetase and its mutants: a study on the molecular target for antifilarial drug development. Mol Biochem Parasitol 246:111426
Arfin SM, Simpson DR, Chiang C, Andrulis IL, Hatfield GW (1977) A role for asparaginyl-tRNA in the regulation of asparagine synthetase in a mammalian cell line. Proc Natl Acad Sci 74:2367–2369. https://doi.org/10.1073/pnas.74.6.2367
Sukuru SCK, Crepin T, Milev Y, Marsh LC, Hill JB, Anderson RJ, Morris JC, Rohatgi A, O’Mahony G, Grøtli M (2006) Discovering new classes of Brugia malayi asparaginyl-tRNA synthetase inhibitors and relating specificity to conformational change. J Comput Aided Mol Des 20:159–178. https://doi.org/10.1007/s10822-006-9043-5
Crepin T, Peterson F, Haertlein M, Jensen D, Wang C, Cusack S, Kron M (2011) A hybrid structural model of the complete Brugia malayi cytoplasmic asparaginyl-tRNA synthetase. J Mol Biol 405:1056–1069. https://doi.org/10.1016/j.jmb.2010.11.049
Dushenkov V, Raskin I (2008) New strategy for the search of natural biologically active substances. Russ J Plant Physiol 55:564–567. https://doi.org/10.1134/S1021443708040201
Süntar I (2020) Importance of ethnopharmacological studies in drug discovery: role of medicinal plants. Phytochem Rev 19:1199–1209. https://doi.org/10.1007/s11101-019-09629-9
Singh S, Singh DB, Singh S, Shukla R, Ramteke PW, Misra K (2019) Exploring medicinal plant legacy for drug discovery in post-genomic era. Biol Sci 89:1141–1151. https://doi.org/10.1007/s40011-018-1013-x
Vishwanathan A, Basavaraju R (2010) A review on Vitex negundo L: A medicinally important plant. Eur J Biol Sci 3:30–42
Basri F, Sharma H, Firdaus S, Jain P, Ranjan A (2014) A review of ethnomedicinal plant-Vitex negundo Linn. Int J Adv Res 2:882–894
Zheng CJ, Li HQ, Ren SC, Xu CL, Rahman K, Qin LP, Sun YH (2015) Phytochemical and pharmacological profile of Vitex negundo. Phytother Res 29:633–647. https://doi.org/10.1002/ptr.5303
Ladda P, Magdum C (2012) Vitex negundo L: Ethnobotany, phytochemistry and pharmacology-a review. Int J Adv Pharm Biol Chem 1:111–120
Behera DR, Bhatnagar S (2018) Filariasis: role of medicinal plant in lymphatic filariasis. Int J Herb Med 6:40–46
Babu S, Nutman TB (2014) Immunology of lymphatic filariasis. Parasite Immunol 36:338–346. https://doi.org/10.1111/pim.12081
Hotterbeekx A, Perneel J, Vieri MK, Colebunders R, Kumar-Singh S (2021) The secretome of filarial nematodes and its role in host-parasite interactions and pathogenicity in onchocerciasis-associated epilepsy. Front Cell Infect Microbiol 11:662766. https://doi.org/10.3389/fcimb.2021.662766
Dhanasekaran JJ, Solaiyappan S, Dhanraj DM, Chatterjee S, Kron M, Dhanasekaran A (2014) Expression and characterisation of asparagine-tRNA synthetase: the key enzyme that mediates angiogenesis in lymphatic filariasis (585.10). FASEB J 28:585510. https://doi.org/10.1096/fasebj.28.1_supplement.585.10
Rupani R, Chavez A (2018) Medicinal plants with traditional use: Ethnobotany in the Indian subcontinent. Clin Dermatol 36:306–309. https://doi.org/10.1016/j.clindermatol.2018.03.005
Alamri MA (2020) Pharmacoinformatics and molecular dynamic simulation studies to identify potential small-molecule inhibitors of WNK-SPAK/OSR1 signaling that mimic the RFQV motifs of WNK kinases. Arab J Chem 13:5107–5117. https://doi.org/10.1016/j.arabjc.2020.02.010
Hollingsworth SA, Dror RO (2018) Molecular dynamics simulation for all. Neuron 99:1129–1143. https://doi.org/10.1016/j.neuron.2018.08.011
Mali SN, Thorat BR, Gupta DR, Pandey A (2021) Mini-review of the importance of hydrazides and their derivatives—synthesis and biological activity. Eng Proc 11:21. https://doi.org/10.3390/ASEC2021-11157
Mohanraj K, Karthikeyan BS, Vivek-Ananth R, Chand R, Aparna S, Mangalapandi P, Samal A (2018) IMPPAT: a curated database of Indian medicinal plants, phytochemistry and therapeutics. Sci Rep 8:1–17. https://doi.org/10.1038/s41598-018-22631-z
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28:235–242. https://doi.org/10.1093/nar/28.1.235
Kaplan W, Littlejohn TG (2001) Swiss-PDB viewer (deep view). Brief Bioinform 2:195–197. https://doi.org/10.1093/bib/2.2.195
Abasian Z, Rostamzadeh A, Mohammadi M, Hosseini M, Rafieian-Kopaei M (2018) A review on role of medicinal plants in polycystic ovarian syndrome: pathophysiology, neuroendocrine signaling, therapeutic status and future prospects. Middle East Fertil Soc J 23:255–262. https://doi.org/10.1016/j.mefs.2018.04.005
Jendele L, Krivak R, Skoda P, Novotny M, Hoksza D (2019) PrankWeb: a web server for ligand binding site prediction and visualization. Nucleic Acids Res 47:W345–W349. https://doi.org/10.1093/nar/gkz424
Dallakyan S, Olson AJ (2015) Small-molecule library screening by docking with PyRx. In Chemical biology; Methods and Protocols, Springer Protocols, pp 243–250. https://doi.org/10.1007/978-1-4939-2269-7_19
Kalimuthu AK, Panneerselvam T, Pavadai P, Pandian SRK, Sundar K, Murugesan S, Ammunje DN, Kumar S, Arunachalam S, Kunjiappan S (2021) Pharmacoinformatics-based investigation of bioactive compounds of Rasam (South Indian recipe) against human cancer. Sci Rep 11:1–19. https://doi.org/10.1038/s41598-021-01008-9
Pires DE, Blundell TL, Ascher DB (2015) pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem 58:4066–4072. https://doi.org/10.1021/acs.jmedchem.5b00104
Kshatriya R, Shelke P, Mali S, Yashwantrao G, Pratap A, Saha S (2021) Synthesis and evaluation of anticancer activity of pyrazolone appended triarylmethanes (TRAMs). ChemistrySelect 6:6230–6239. https://doi.org/10.1002/slct.202101083
Mvondo JGM, Matondo A, Mawete DT, Bambi S-MN, Mbala BM, Lohohola PO (2021) In silico ADME/T properties of quinine derivatives using SwissADME and pkCSM webservers. Int J Trop Dis Health 42:1–12. https://doi.org/10.9734/ijtdh/2021/v42i1130492
Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ (2005) GROMACS: fast, flexible, and free. J Comput Chem 26:1701–1718. https://doi.org/10.1002/jcc.20291
Desale VJ, Mali SN, Thorat BR, Yamgar RS (2021) Synthesis, admetSAR predictions, DPPH radical scavenging activity, and potent anti-mycobacterial studies of hydrazones of substituted 4-(anilino methyl) benzohydrazides (Part 2). Curr Comput Aided Drug Des 17:493–503. https://doi.org/10.2174/1573409916666200615141047
Van Aalten DM, Bywater R, Findlay JB, Hendlich M, Hooft RW, Vriend G (1996) PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. J Comput Aided Mol Des 10:255–262. https://doi.org/10.1007/BF00355047
Rajeshkumar RR, Kumar BK, Parasuraman P, Panneerselvam T, Sundar K, Ammunje DN, Pandian SRK, Murugesan S, Kabilan SJ, Kunjiappan S (2022) Graph theoretical network analysis, in silico exploration, and validation of bioactive compounds from Cynodon dactylon as potential neuroprotective agents against α-synuclein. BioImpacts 12:487–499. https://doi.org/10.34172/bi.2022.24113
Palanichamy C, Pavadai P, Panneerselvam T, Arunachalam S, Babkiewicz E, Ram Kumar Pandian S, ShanmugampillaiJeyarajaguru K, NayakAmmunje D, Kannan S, Chandrasekaran J (2022) Aphrodisiac performance of bioactive compounds from Mimosa pudica Linn: in silico molecular docking and dynamics simulation approach. Molecules 27:3799. https://doi.org/10.3390/molecules27123799
Acknowledgements
The authors acknowledge the Department of Biotechnology, Kalasalingam Academy of Research and Education, Krishnankoil, Tamil Nadu, India, for providing essential facilities to perform the research work, and Department of Pharmaceutical Chemistry, Faculty of Pharmacy, M.S. Ramaiah University of Applied Sciences, Bengaluru, for providing computational facility for research work.
Funding
Funding acquisition from Department of Biotechnology, Kalasalingam Academy of Research and Education, India.
Author information
Authors and Affiliations
Contributions
Shanmugampillai Jeyarajaguru Kabilan: conceptualization, reviewing and editing, resources, and supervision.
Selvaraj Kunjiappan: simulations, data collection, analysis, and writing—original draft preparation.
Krishnan Sundar: reviewing and editing, resources, and formal analysis.
Parasuraman Pavadai: simulations, data collection, analysis, and writing—original draft preparation.
Nivethitha Sathishkumar: material preparation, data collection, analysis, and writing—original draft preparation.
Haritha Velayuthaperumal: material preparation, data collection, analysis, and writing—original draft preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
We approved all ethics.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kabilan, S.J., Kunjiappan, S., Sundar, K. et al. Pharmacoinformatics-based screening of active compounds from Vitex negundo against lymphatic filariasis by targeting asparaginyl-tRNA synthetase. J Mol Model 29, 87 (2023). https://doi.org/10.1007/s00894-023-05488-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-023-05488-6