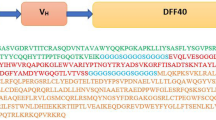
Abstract
Breast cancer remains the most frequently diagnosed cancer and the principal cause of mortality by malignancy in women. HER2 positive subtype includes 15–20% of breast cancer cases. This receptor could be an appropriate mark for targeting breast cancer cells. Immunotherapy methods compared to current cancer treatment methods have the lowest side effects. DELTA-stichotoxin-Hmg2a is isolated from the sea anemone and kills cells through pore formation. In the current study, we designed and evaluated an immunotoxin composed of pertuzumab and DELTA-stichotoxin-Hmg2a-derived scFv by bioinformatics tools. The designed immunotoxin was constructed using the amino acid sequences. Then, secondary structure and physico-chemical features were studied, and the tertiary structure of the immunotoxin was built according to the homology modeling methods. The validation and allergenicity of the model were assessed. The immunotoxin and receptor were docked and molecular dynamics simulation indicated the construct stability. The analysis results indicated that the construct is a stable protein that could have a natural-like structure and would not be an allergen, so this immunotoxin could effectively target HER2 receptors. Therefore, our designed immunotoxin could be an appropriate immunotoxin against HER2-positive breast cancer and could be a challenging topic for future in vitro and in vivo studies.













Similar content being viewed by others
Data availability
N/A.
Code availability
N/A.
References
Britt KL, Cuzick J, Phillips K-A (2020) Key steps for effective breast cancer prevention. Nat Rev Cancer 20(8):417–436
Mercogliano MF, Bruni S, Elizalde PV, Schillaci R (2020) Tumor necrosis factor α blockade: an opportunity to tackle breast Cancer. Front Oncol 10:584
Turashvili G, Brogi E (2017) Tumor heterogeneity in breast cancer. Front Med 4:227
Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM et al (2012) Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 486(7403):405–409
Makena MR, Rao R (2020) Subtype specific targeting of calcium signaling in breast cancer. Cell Calcium 85:102109
Massicano AV, Marquez-Nostra BV, Lapi SE (2018) Targeting HER2 in nuclear medicine for imaging and therapy. Mol Imaging 17:1536012117745386
Lavaud P, Rousseau B, Ajgal Z, Arrondeau J, Huillard O, Alexandre J et al (2016) Bi-weekly very-high-dose lapatinib: an easy-to-use active option in HER-2-positive breast cancer patients with meningeal carcinomatosis. Breast Cancer Res Treat 157(1):191
Dong Y, Li W, Gu Z, Xing R, Ma Y, Zhang Q et al (2019) Inhibition of HER2-positive breast cancer growth by blocking the HER2 signaling pathway with HER2-glycan-imprinted nanoparticles. Angew Chem Int Ed 58(31):10621–10625
Recondo G Jr, de la Vega M, Galanternik F, Díaz-Cantón E, Leone BA, Leone JP (2016) Novel approaches to target HER2-positive breast cancer: trastuzumab emtansine. Cancer Manag Res 8:57
Kim HJ, Park JY, Lee TS, Song IH, Cho YL, Chae JR et al (2019) PET imaging of HER2 expression with an 18F-fluoride labeled aptamer. PLoS ONE 14(1):e0211047
Ernst B, Anderson KS (2015) Immunotherapy for the treatment of breast cancer. Curr Oncol Rep 17(2):5
Boyraz B, Sendur MA, Aksoy S, Babacan T, Roach EC, Kizilarslanoglu MC et al (2013) Trastuzumab emtansine (T-DM1) for HER2-positive breast cancer. Curr Med Res Opin 29(4):405–414
Capelan M, Pugliano L, De Azambuja E, Bozovic I, Saini K, Sotiriou C et al (2013) Pertuzumab: new hope for patients with HER2-positive breast cancer. Ann Oncol 24(2):273–282
Skerra A, Pluckthun A (1988) Assembly of a functional immunoglobulin Fv fragment in Escherichia coli. Science 240(4855):1038–1041
Kato M, Hanyu Y (2013) Construction of an scFv library by enzymatic assembly of VL and VH genes. J Immunol Methods 396(1–2):15–22
Vallet-Courbin A, Larivière M, Hocquellet A, Hemadou A, Parimala S-N, Laroche-Traineau J et al (2017) A recombinant human anti-platelet SCFV antibody produced in pichia pastoris for atheroma targeting. PLoS ONE 12(1):e0170305
Bustamante-Córdova L, Melgoza-González EA, Hernández J (2018) Recombinant antibodies in veterinary medicine: an update. Front Vet Sci 5:175
Hull EA (2015) Antibody conjugates via disulfide bridging: towards therapeutic and diagnostic applications: UCL (University College London)
Rezaie E, Pour AB, Amani J, Hosseini HM (2020) Bioinformatics predictions, expression, purification and structural analysis of the PE38KDEL-scfv immunotoxin against EPHA2 receptor. Int J Pept Res Ther 26(2):979–996
Weigel KJ, Shen L, Thomas CL, Alber D, Drapalik L, Schafer ZT et al (2015) Design and evaluation of a peptide-based immunotoxin for breast cancer therapeutics. FEBS Open Bio 5:202–208
Ramezanpour M, Da Silva KB, Sanderson BJ (2014) The effect of sea anemone (H. magnifica) venom on two human breast cancer lines: death by apoptosis. Cytotechnology 66(5):845–52
Madio B, Peigneur S, Chin YK, Hamilton BR, Henriques ST, Smith JJ et al (2018) PHAB toxins: a unique family of predatory sea anemone toxins evolving via intra-gene concerted evolution defines a new peptide fold. Cell Mol Life Sci 75(24):4511–4524
Wang Y, Chua KL, Khoo HE (2000) A new cytolysin from the sea anemone, Heteractis magnifica: isolation, cDNA cloning and functional expression. Biochimica et Biophysica Acta (BBA)-Protein Structure and Molecular Enzymology 1478(1):9–18
Madio B, King GF, Undheim EA (2019) Sea anemone toxins: a structural overview. Mar Drugs 17(6):325
Ker D-S, Sha HX, Jonet MA, Hwang JS, Ng CL (2021) Structural and functional analysis of Hydra Actinoporin-Like Toxin 1 (HALT-1). Sci Rep 11(1):1–13
Subburaj Y, Ros U, Hermann E, Tong R, García-Sáez AJ (2015) Toxicity of an α-pore-forming toxin depends on the assembly mechanism on the target membrane as revealed by single molecule imaging. J Biol Chem 290(8):4856–4865
Rojko N, Dalla Serra M, Maček P, Anderluh G (2016) Pore formation by actinoporins, cytolysins from sea anemones. Biochimica et Biophysica Acta (BBA)-Biomembranes 1858(3):446-56
Ramírez-Carreto S, Miranda-Zaragoza B, Rodríguez-Almazán C (2020) Actinoporins: from the structure and function to the generation of biotechnological and therapeutic tools. Biomolecules 10(4):539
Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch AJTpph (2005) Protein identification and analysis tools on the ExPASy server. 571–607
Dimitrov I, Naneva L, Doytchinova I, Bangov I (2014) AllergenFP: allergenicity prediction by descriptor fingerprints. Bioinformatics 30(6):846–851
Dimitrov I, Flower DR, Doytchinova I, editors (2013) AllerTOP-a server for in silico prediction of allergens. BMC bioinformatics. BioMed Central
Yang J, Zhang Y (2015) I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res 43(W1):W174–W181
Heo L, Park H, Seok C (2013) GalaxyRefine: protein structure refinement driven by side-chain repacking. Nucleic Acids Res 41(W1):W384–W388
Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35(suppl_2):W407-W10
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures 26(2):283–91
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2(9):1511–1519
Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C et al (2017) The ClusPro web server for protein–protein docking. Nat Protoc 12(2):255
Cao W, Chen H-D, Yu Y-W, Li N, Chen W-Q (2021) Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J 134(07):783–791
Zhai Z, Zhang F, Zheng Y, Zhou L, Tian T, Lin S et al (2019) Effects of marital status on breast cancer survival by age, race, and hormone receptor status: a population-based Study. Cancer Med 8(10):4906–4917
Wang J, Xu B (2019) Targeted therapeutic options and future perspectives for HER2-positive breast cancer. Signal Transduct Target Ther 4(1):1–22
Lima ZS, Ghadamzadeh M, Arashloo FT, Amjad G, Ebadi MR, Younesi L (2019) Recent advances of therapeutic targets based on the molecular signature in breast cancer: genetic mutations and implications for current treatment paradigms. J Hematol Oncol 12(1):1–25
Yan Y, Zhang L, Zuo Y, Qian H, Liu C (2020) Immune checkpoint blockade in cancer immunotherapy: mechanisms, clinical outcomes, and safety profiles of PD-1/PD-L1 inhibitors. Arch Immunol Ther Exp 68(6):1–15
Jagosky M, Tan AR (2021) Combination of pertuzumab and trastuzumab in the treatment of HER2-positive early breast cancer: a review of the emerging clinical data. Breast Cancer 13:393
Ayoub NM, Al-Shami KM, Yaghan RJ (2019) Immunotherapy for HER2-positive breast cancer: recent advances and combination therapeutic approaches. Breast Cancer 11:53
HashemiYeganeh H, Heiat M, Kieliszek M, Alavian SM, Rezaie E (2021) DT389-YP7, a recombinant immunotoxin against glypican-3 that inhibits hepatocellular cancer cells: an in vitro study. Toxins 13(11):749
Tung BT, Quynh CTX, Hong NK (2022) Nanotechnology applications in breast cancer. Handbook of Research on Natural Products and Their Bioactive Compounds as Cancer Therapeutics: IGI Global. 442–65
Mahmoudi R, Dianat-Moghadam H, Poorebrahim M, Siapoush S, Poortahmasebi V, Salahlou R et al (2021) Recombinant immunotoxins development for HER2-based targeted cancer therapies. Cancer Cell Int 21(1):1–17
Heiat M, HashemiYeganeh H, Alavian SM, Rezaie E (2021) Immunotoxins immunotherapy against hepatocellular carcinoma: a promising prospect. Toxins 13(10):719
Rezaie E, Bidmeshki Pour A, Amani J, Mahmoodzadeh HH (2020) Bioinformatics predictions, expression, purification and structural analysis of the PE38KDEL-scfv immunotoxin against EPHA2 receptor. Int J Pept Res Ther 26(2):979–996
Rong L, Lim RM, Yin X, Tan L, Yang JH, Xie J (2022) Site-specific dinitrophenylation of single-chain antibody fragments for redirecting a universal CAR-T cell against cancer antigens. J Mol Biol 434(8):167513
Krokhotin A, Du H, Hirabayashi K, Popov K, Kurokawa T, Wan X et al (2019) Computationally guided design of single-chain variable fragment improves specificity of chimeric antigen receptors. Mol Ther Oncolytics 15:30–37
Cho H-S, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, et al (2003) Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. 421(6924):756–60
Funding
This study was financially supported by the Behbahan Faculty of Medical Sciences, Behbahan, Iran (grant number: 400055).
Author information
Authors and Affiliations
Contributions
The authors contributed as below:
Writing: Zeinab Ghesmati, Samira Mokhtari, and Maliheh Parvanak; data analysis: Mortaza Taheri-Anganeh, Khadijeh Ahmadi, and Farzaneh Vahedi; critical editing: Vahid Zarezade and Zeinab Shajirat; supervising and conceptualization: Navid Nezafat and Ahmad Movahedpour. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ghesmati, Z., Mokhtari, S., Parvanak, M. et al. Designing a humanized immunotoxin based on DELTA-stichotoxin-Hmg2a toxin: an in silico study. J Mol Model 28, 392 (2022). https://doi.org/10.1007/s00894-022-05389-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-022-05389-0