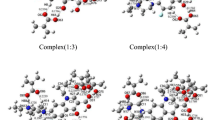
Abstract
Molecularly imprinted polymers (MIP) are the polymers created by molecular imprinting techniques that leave cavities for the specific interactions with a template molecule and have been applied in molecular selectivity tasks. In this study, the molecular dynamics (MD) simulation technique was used to demonstrate that aniline oligomer could be developed as a potential MIP for detection and separation of the spectinomycin drug molecule for gonorrhea treatment. MD simulations were performed for the systems of a spectinomycin within aniline oligomers of different sizes. The mean square displacement (MSD) and the diffusivity calculated from MD simulations showed that the diffusion coefficient was significantly dropped when the length of aniline oligomer was greater than two. The diffusion coefficient of spectinomycin became the lowest within aniline trimers, corresponded to the highest atomic distribution of MIP around the template. Then, the specific cavity in MIP systems with and without spectinomycin was calculated to assess the stability of the cavity created by the template. The volume of a cavity created within the trimer system was closest to the spectinomycin volume and therefore became the optimal oligomer size for further development of MIP.
Graphical Abstract






Similar content being viewed by others
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary material.
Code availability
Not applicable.
References
Gornik T, Shinde S, Lamovsek L, Koblar M, Heath E, Sellergren B, Kosjek T (2020) Molecularly imprinted polymers for the removal of antide-pressants from contaminated wastewater. Polymers 13(1):120
Krupadam RJ, Patel GP, Balasubramanian R (2011) Removal of cyanotoxins from surface water resources using reusable molecularly imprinted polymer adsorbents. Environ Sci Pollut Res 19(5):1841–1851
Lee W-C, Cheng C-H, Pan H-H, Chung T-H, Hwang C-C (2007) Chromatographic characterization of molecularly imprinted polymers. Anal Bioanal Chem 390(4):1101–1109
Tóth B, László K, Horvai G (2005) Chromatographic behavior of silica–polymer composite molecularly imprinted materials. J Chromatogr A 1100(1):60–67
Yang S, Wang Y, Jiang Y, Li S, Liu W (2016) Molecularly imprinted polymers for the identification and separation of chiral drugs and biomolecules. Polymers 8(6):216
Shrivastav AM, Usha SP, Gupta BD (2017) Highly sensitive and selective erythromycin nanosensor employing fiber optic SPR/ERY imprinted nanostructure: application in milk and honey. Biosens Bioelectron 90:516–524
Shimizu KD, Stephenson CJ (2010) Molecularly imprinted polymer sensor arrays. Curr Opin Chem Biol 14(6):743–750
Ansell RJ, Mosbach K (1998) Magnetic molecularly imprinted polymer beads for drug radioligand binding assay. Analyst 123(7):1611–1616
Hiratani H, Mizutani Y, Alvarez-Lorenzo C (2005) Controlling drug release from imprinted hydrogels by modifying the characteristics of the imprinted cavities. Macromol Biosci 5(8):728–733
Luliński P (2013) Molecularly imprinted polymers as the future drug delivery devices. Acta Poloniae Pharmaceutica 70(4):601–609
Sharma G, Kandasubramanian B (2020) Molecularly imprinted polymers for selective recognition and extraction of heavy metal ions and toxic dyes. Journal of Chemical & Engineering Data 65(2):396–418
Bagdžiūnas G (2020) Theoretical design of molecularly imprinted polymers based on polyaniline and polypyrrole for detection of tryptophan. Mol Syst Design Eng 5(9):1504–1512
Zhang B, Fan X, Zhao D (2018) Computer-aided design of molecularly imprinted polymers for simultaneous detection of clenbuterol and its metabolites. Polymers 11(1):17
Wungu T D K, Marsha S E, Widayani, Suprijadi (2017) Density functional theory (DFT) study of molecularly imprinted polymer (MIP) methacrylic acid (MAA) with D-glucose. IOP Conference Series Materials Science and Engineering, 214(1):012004
Malde AK, Zuo L, Breeze M, Stroet M, Poger D, Nair PC, Oostenbrink C, Mark AE (2011) An automated force field Topology Builder (ATB) and repository. Chem Theory Comput 7:4026–4037
Luo D, Zhao Z, Zhang L, Wang Q, Wang J (2013) On the structure of molecularly imprinted polymers by modifying charge on functional groups through molecular dynamics simulations. Mol Simul 40(6):431–438
Monte AD, Laffi G, Mancini G (1994) Occupational contact dermatitis due to spectinomycin. Contact Dermatitis 31(3):204–205
Chu T X, Vu VP, Tran H T, Tran T L, Tran Q T, Manh T L (2020) Molecularly imprinted polyaniline nanowire-based electrochemical biosensor for chloramphenicol detection: A kinetic study of aniline electropolymerization. Journal of The Electrochemical Society 167(2):027527
Roy AK, Nisha SV, Dhand C, Malhotra BD (2011) Molecularly imprinted polyaniline film for ascorbic acid detection. J Mol Recog 24(4):700–706
Bagdžiūnas G (2020) Theoretical design of molecularly imprinted polymers based on polyaniline and polypyrrole for detection of tryptophan. Mol Syst Design Eng 5(9):1504–1515
Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, Wang J, Yu B, Zhang J, Bryant SH (2016) PubChem substance and compound databases. Nucleic Acids Res 44(D1):D1202–D1213
Hanwell M D, Curtis D E, Lonie D C, Vandermeersch T, Zurek E, Hutchison G R (2012) Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. Journal of Cheminformatics 4(17):1–17
Schmid N, Eichenberger AP, Choutko A, Riniker S, Winger M, Mark AE, Van Gunsteren WF (2011) Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur Biophys J 40(7):843–856
Zhao Z-J, Wang Q, Zhang L, Wu T (2008) Structured water and water−polymer interactions in hydrogels of molecularly imprinted polymers. J Phys Chem B 112(25):7515–7521
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19–25
Wallace AC, Laskowski RA, Thornton JM (1995) LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng 8:127–134
He Y, Hu T, Zeng D (2019) Scan-flood Fill(SCAFF): an efficient automatic precise region filling algorithm for complicated regions. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops 1:761–769
Piletsky SA, Guerreiro A, Piletska EV, Chianella I, Karim K, Turner APF (2004) Polymer Cookery 2 Influence of polymerization pressure and polymer swelling on the performance of molecularly imprinted polymers. Macromolecules 37(13):5018–5022
Funding
The authors are grateful for the research funding provided by the Department of Physics and the Center of Excellence in Theoretical and Computational Science (TaCS), KMUTT.
Author information
Authors and Affiliations
Contributions
Chanadan Douykhumklaw performed all the simulations and the drafts of the manuscript under the supervision of Thana Sutthibutpong.
Corresponding author
Ethics declarations
Conflict of interest
All authors have participated in (a) conception and design or analysis and interpretation of the data, (b) drafting the article or revising it critically for important intellectual content, and (c) approval of the final version. This manuscript has not been submitted to, nor is under review at, another journal or other publishing venue. The authors have no affiliation with any organization with a direct or indirect financial interest in the subject matter discussed in the manuscript. The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Douykhumklaw, C., Sutthibutpong, T. A molecular dynamics study on the diffusion and imprint ability of spectinomycin under different sizes of aniline oligomers. J Mol Model 28, 387 (2022). https://doi.org/10.1007/s00894-022-05371-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-022-05371-w