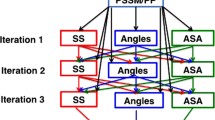
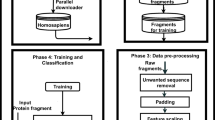
Abstract
Proteins are constructed from amino acid sequences. Their structural classifications include primary, secondary, tertiary, and quaternary, with tertiary and quaternary structures influencing protein function. Because a protein’s structure is inextricably connected to its biological function, machine learning algorithms that can better anticipate the structures have the potential to lead to new scientific discoveries in human health and improve our capacity to develop new treatments. Protein secondary structure assignment enriches the structural and functional understanding of proteins. It helps in protein structure comparison and classification studies, besides facilitating secondary and tertiary structure prediction systems. Several secondary structure assignment methods have been developed since the 1980s, most of which are based on hydrogen bond analysis and atomic coordinate features. However, the assignment process becomes complex when protein data includes missing atoms. Deep neural networks are often referred to as universal function approximators because they can approximate any function to produce the desired output when properly designed and trained. Optimised deep learning architectures have already proven their ability to increase performance in a wide range of problems. Recently, the ResNet architecture has garnered significant interest due to its applicability in various areas, including image classification and protein contact map prediction. The proposed model, which is based on the ResNet architecture, assigns secondary structures using Cα atom coordinates. The model achieved an accuracy of 94% when evaluated against the benchmark and independent test sets. The findings encourage the development of new deep learning-based methods that are more generalised across various protein learning tasks. Furthermore, it allows computational biologists to delve deeper into integrating these techniques with experimental methods. The model codes are available at: https://github.com/jisnava/ResNet_for_Structure_Assignments/.










Similar content being viewed by others
Data availability
The data is available at: https://github.com/jisnava/ResNet_for_Structure_Assignments/.
Code availability
The model codes are made open at: https://github.com/jisnava/ResNet_for_Structure_Assignments/.
References
Pauling L, Corey RB, Branson HR (1951) The structure of proteins: two hydrogen-bonded helical configurations of the polypeptide chain. Proc Natl Acad Sci 37(4):205–211
Andersen CA, Rost B (2003) Secondary structure assignment. Methods Biochem Anal 44:341–364
Andersen CA, Rost B (2009) Secondary structure assignment. Structural Bioinformatics 44:459–484
Murzin AG, Brenner SE, Hubbard T, Chothia C (1995) Scop: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol 247(4):536–540
Sayle RA, Milner-White EJ (1995) Rasmol: biomolecular graphics for all. Trends Biochem Sci 20(9):374–376
Fischel-Ghodsian F, Mathiowitz G, Smith TF (1990) Alignment of protein sequences using secondary structure: a modified dynamic programming method. Protein Eng Des Sel 3(7):577–581
Fischer D, Eisenberg D (1996) Protein fold recognition using sequence-derived predictions. Protein Sci 5(5):947–955
A. Fiser (2010), Template-based protein structure modeling, in: Computational biology, Springer, 73–94.
Torrisi M, Kaleel M, Pollastri G (2019) Deeper profiles and cascaded recurrent and convolutional neural networks for state-of-the-art protein secondary structure prediction. Sci Rep 9(1):1–12
W. Kabsch, C. Sander (1983), Dictionary of protein secondary structure:pattern recognition of hydrogen-bonded and geometrical features, Biopolymers: Original Research on Biomolecules 22 (12) 2577–2637.
King SM, Johnson WC (1999) Assigning secondary structure from protein coordinate data, Proteins: Structure. Function, and Bioinformatics 35(3):313–320
Cubellis MV, Cailliez F, Lovell SC (2005) Secondary structure assignment that accurately reflects physical and evolutionary characteristics. BMC Bioinformatics 6(4):1–9
F Dupuis, J-F Sadoc, J-P Mornon (2004) Protein secondary structure assignment through voronoi tessellation, Proteins: structure, function, and bioinformatics 55 (3) 519–528
Zhang W, Dunker AK, Zhou Y (2008) Assessing secondary structure assignment of protein structures by using pairwise sequence-alignment benchmarks, Proteins: Structure. Function, and Bioinformatics 71(1):61–67
Park S-Y, Yoo M-J, Shin J-M, Cho K-H (2011) Saba (secondary structure assignment program based on only alpha carbons): a novel pseudo center geometrical criterion for accurate assignment of protein secondary structures. BMB Rep 44(2):118–122
Heinig M, Frishman D (2004) STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res 32(suppl2):W500–W502
Adasme-Carre ̃no F, Caballero J, Ireta J (2021) Psique: protein secondary structure identification on the basis of quaternions and electronic structure calculations. J Chem Inf Model 61(4):1789–1800
Brinkjost T, Ehrt C, Koch O, Mutzel P (2020) Scot: rethinking the classification of secondary structure elements. Bioinformatics 36(8):2417–2428
Kumar P, Bansal M (2015) Identification of local variations within secondary structures of proteins. Acta Crystallogr D Biol Crystallogr 71(5):1077–1086
Labesse G, N. Colloc’h, J. Pothier, J.-P. Mornon, (1997) P-sea: a new efficient assignment of secondary structure from cα trace of proteins. Bioinformatics 13(3):291–295
Koch O, Cole J (2011) An automated method for consistent helix assignment using turn information, Proteins: Structure. Function, and Bioinformatics 79(5):1416–1426
Srinivasan R, Rose GD (1999) A physical basis for protein secondary structure. Proc Natl Acad Sci 96(25):14258–14263
Fodje M, Al-Karadaghi S (2002) Occurrence, conformational features and amino acid propensities for the π-helix. Protein Eng Des Sel 15(5):353–358
Nagy G, Oostenbrink C (2014) Dihedral-based segment identification and classification of biopolymers i: proteins. J Chem Inf Model 54(1):266–277
Hosseini S-R, Sadeghi M, Pezeshk H, Eslahchi C, Habibi M (2008) Prosign: a method for protein secondary structure assignment based on three-dimensional coordinates of consecutive cα atoms. Comput Biol Chem 32(6):406–411
Majumdar I, Krishna SS, Grishin NV (2005) Palsse: a program to delineate linear secondary structural elements from protein structures. BMC Bioinformatics 6(1):202
Taylor WR (2001) Defining linear segments in protein structure. J Mol Biol 310(5):1135–1150
Martin J, Letellier G, Marin A, Taly J-F, de Brevern AG, Gibrat J-F (2005) Protein secondary structure assignment revisited: a detailed analysis of different assignment methods. BMC Struct Biol 5(1):17
Cao C, Wang G, Liu A, Xu S, Wang L, Zou S (2016) A new secondary structure assignment algorithm using cαbackbone fragments. Int J Mol Sci 17(3):333
Jordan MI, Mitchell TM (2015) Machine learning: trends, perspectives, and prospects. Science 349(6245):255–260
Wu Y, Ianakiev K, Govindaraju V (2002) Improved k-nearest neighbor classification. Pattern Recognit 35(10):2311–2318. https://doi.org/10.1016/S0031-3203(01)00132-7
Law SM, Frank AT, Brooks CL III (2014) Pcasso: a fast and efficient cα-based method for accurately assigning protein secondary structure elements. J Comput Chem 35(24):1757–1761
Salawu EO (2016) Rafosa: random forests secondary structure assignment for coarse-grained and all-atom protein systems. Cogent Biology 2(1):1214061
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436–444
Goh GB, Hodas NO, Vishnu A (2017) Deep learning for computational chemistry. J Comput Chem 38(16):1291–1307
Jisna VA, Jayaraj PB (2021) Protein structure prediction: conventional and deep learning perspectives. Protein J 40(4):522–544
Antony JV, Madhu P, Balakrishnan JP, Yadav H (2021) Assigning secondary structure in proteins using ai. J Mol Model 27(9):1–13
Wang, L, Cao C, Zuo S (2021) Protein secondary structure assignment using pc‐polyline and convolutional neural network. Proteins: Structure, Function, and Bioinformatics 89(8):1017–1029
Wang G, Dunbrack RL (2005) Pisces: recent improvements to a pdb sequence culling server. Nucleic Acids Res 33(suppl2):W94–W98
Rose PW, Bi C, Bluhm WF, Christie CH, Dimitropoulos D, Dutta S, Green RK, Goodsell DS, Prli ́c A, Quesada M et al (2012) The rcsb protein data bank: new resources for research and education. Nucleic Acids Res 41(D1):D475–D482
Hochreiter S, Schmidhuber J (1997) Long short-term memory. Neural Comput 9(8):1735–1780
Werbos PJ (1990) Backpropagation through time: what it does and how to do it. Proc IEEE 78(10):1550–1560
Hecht-Nielsen R (1992) Theory of the backpropagation neural network. Neural networks for perception. Academic Press, pp 65–93
Sazli MH (2006) A brief review of feed-forward neural networks. Communications Faculty of Sciences University of Ankara Series A2-A3 Physical Sciences and Engineering 50(01)
Bengio Y, Simard P, Frasconi P (1994) Learning long-term dependencies with gradient descent is difficult. IEEE Trans Neural Networks 5(2):157–166
Zeiler MD, Ranzato D, Monga R, Mao M, Yang K, Le QV, Nguyen P et al ( 2013) On rectified linear units for speech processing. In: 2013 IEEE International Conference on Acoustics, Speech and Signal Processing. IEEE, pp 3517–3521
Wu Z, Chunhua S, Van Den Hengel A (2019) Wider or deeper: Revisiting the resnet model for visual recognition. Pattern Recognit 90:119–133
Yamashita R, Nishio M, Do RKG, Togashi K (2018) Convolutional neural networks: an overview and application in radiology. Insights Imaging 9(4):611–629
Kim P (2017) Convolutional neural network. In: MATLAB deep learning. Apress, Berkeley, pp 121–147
Sermanet P, Chintala S, LeCun Y (2012) November), Convolutional neural networks applied to house numbers digit classification, In Proceedings of the 21st international conference on pattern recognition (ICPR2012) ( 3288–3291) IEEE.
Krizhevsky A, Sutskever I, Hinton GE (2017) Imagenet classification with deep convolutional neural networks. Commun ACM 60(6):84–90
R Pascanu, T Mikolov, Y Bengio (2013) On the difficulty of training recurrent neural networks, In: International conference on machine learning, PMLR, 1310–1318.
Glorot X, Bordes A, Bengio Y (2011) Deep sparse rectifier neural networks. In: Proceedings of the fourteenth international conference on artificial intelligence and statistics. JMLR Workshop and Conference Proceedings, pp 315–323
Ioffe S, Szegedy C (2015 June) Batch normalization: accelerating deep network training by reducing internal covariate shift, In International conference on machine learning ( 448–456) PMLR.
Araujo A, Norris W, Sim J (2019 ) Computing receptive fields of convolutional neural networks. Distill 4(11):e21
Zhao Y, Liu Y (2021) Oclstm: optimized convolutional and long short-term memory neural network model for protein secondary structure prediction. PLoS ONE 16(2):e0245982
Heffernan R, Yang Y, Paliwal K, Zhou Y (2017) Capturing non-local interactions by long short-term memory bidirectional recurrent neural networks for improving prediction of protein secondary structure, backbone angles, contact numbers and solvent accessibility. Bioinformatics 33(18):2842–2849
Acknowledgements
The authors thank the Centre for Computational Modelling and Simulation (CCMS) and Central Computer Centre (CCC) at the National Institute of Technology Calicut, for providing the NVIDIA DGX station facility to train the deep neural network architectures.
Funding
It is part of my (V. A. Jisna) PhD work at the National Institute of Technology Calicut, India. The research is funded by the Ministry of Human Resource Development, India.
Author information
Authors and Affiliations
Contributions
Jisna Vellara Antony (JVA) did the conceptualisation and dataset construction. JVA and Roosafeed Koya (RK) implemented the models. Jayaraj Pottekkattuvalappil Balakrishnan (JPB), Pulinthanathu Narayanan Pournami (PNP), and Gopakumar Gopalakrishnan Nair (GGN) supervised the project. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Antony, J.V., Koya, R., Pournami, P.N. et al. Protein secondary structure assignment using residual networks. J Mol Model 28, 269 (2022). https://doi.org/10.1007/s00894-022-05271-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-022-05271-z