Abstract
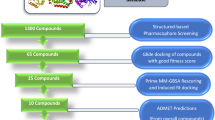
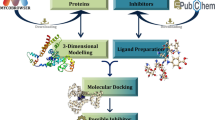
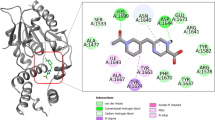
Tuberculosis caused by Mycobacterium tuberculosis (Mtb) is responsible for the highest global health problem, with the deaths of millions of people. With prevalence of multiple drug resistance (MDR) strains and extended therapeutic times, it is important to discover small molecule inhibitors against novel hypothetical proteins of the pathogen. In this study, a virtual screening protocol was carried out against MtbH37Rv hypothetical protein RipD (Rv1566c) for the identification of potential small molecule inhibitors. The 3D model of the protein structure binding site was used for virtual screening (VS) of inhibitors from the Pathogen Box, followed by its validation through a molecular docking study. The stability of the protein–ligand complex was assessed using a 150 ns molecular dynamics simulation. MM-PBSA and MM-GBSA are the two approaches that were used to perform the trajectory analysis and determine the binding free energies, respectively. The ligand binding was observed to be stable across the entire time frame with an approximate binding free energy of -22.9916 kcal/mol. The drug-likeness of the inhibitors along with a potential anti-tuberculosis compound was validated by ADMET prediction software. Furthermore, a CFU inhibition assay was used to validate the best hit compound’s in vitro inhibitory efficacy against a non-pathogenic Mycobacterium smegmatis MC2155 under low nutrient culture conditions. The study reported that the compound proposed in our study (Pathogen Box ID: MMV687700) will be useful for the identification of potential inhibitors against Mtb in future.






Similar content being viewed by others
Data availability
Available on request.
Code availability
N/A.
References
Chakaya J, Khan M, Ntoumi F, Aklillu E, Fatima R, Mwaba P, Kapata N, Mfinanga S, Hasnain SE, Katoto PD, Bulabula AN (2021) Global Tuberculosis Report 2020–Reflections on the Global TB burden, treatment and prevention efforts. Int J Infect Dis 113:S7-12
Yang Z, Zeng X, Tsui SK (2019) Investigating function roles of hypothetical proteins encoded by the Mycobacterium tuberculosis H37Rv genome. BMC Genomics 20(1):1
Marinova D, Gonzalo-Asensio J, Aguilo N, Martin C (2017) MTBVAC from discovery to clinical trials in tuberculosis-endemic countries. Expert Rev Vaccines 16(6):565–576
Ottenhoff THM, Kaufmann SHE (2012) Vaccines against tuberculosis: where are we and where do we need to go? PLoS Pathog 8(5):e1002607
Velayati AA, Masjedi MR, Farnia P, Tabarsi P, Ghanavi J, ZiaZarifi AH et al (2009) Emergence of new forms of totally drug-resistant tuberculosis bacilli: super extensively drug-resistant tuberculosis or totally drug-resistant strains in Iran. Chest 136(2):420–425
Velayati AA, Farnia P, Hoffner S (2018) Drug-resistant Mycobacterium tuberculosis: Epidemiology and role of morphological alterations. J Glob Antimicrob Resist 12:192–196
Uplekar M, Weil D, Lonnroth K, Jaramillo E, Lienhardt C, Dias HM et al (2015) WHO’s new end TB strategy. Lancet 385(9979):1799–1801
Chaturvedi V, Dwivedi N, Tripathi RP, Sinha S (2007) Evaluation of Mycobacterium smegmatis as a possible surrogate screen for selecting molecules active against multi-drug resistant Mycobacterium tuberculosis. J Gen Appl Microbiol 53(6):333–337
Lauzardo M, Peloquin CA (2016) Tuberculosis therapy for 2016 and beyond. Expert Opin Pharmacother 17(14):1859–1872
Divakar DD, Jastaniyah NT, Altamimi HG, Alnakhli YO, Alkheraif AA, Haleem S (2018) Enhanced antimicrobial activity of naturally derived bioactive molecule chitosan conjugated silver nanoparticle against dental implant pathogens. Int J Biol Macromol 108:790–797
Machowski EE, Senzani S, Ealand C, Kana BD (2014) Comparative genomics for mycobacterial peptidoglycan remodelling enzymes reveals extensive genetic multiplicity. BMC Microbiol 14(1):1–2
Catalão MJ, Filipe SR, Pimentel M (2019) Revisiting anti-tuberculosis therapeutic strategies that target the peptidoglycan structure and synthesis. Front Microbiol 10:190
Amir A, Rana K, Arya A, Kapoor N, Kumar H, Siddiqui MA (2014) Mycobacterium tuberculosis H37Rv: in silico drug targets identification by metabolic pathways analysis. Int J Evol Biol 2014:284170
Park Y, Pacitto A, Bayliss T, Cleghorn LAT, Wang Z, Hartman T et al (2017) Essential but not vulnerable: indazole sulfonamides targeting inosine monophosphate dehydrogenase as potential leads against Mycobacterium tuberculosis. ACS Infect Dis 3(1):18–33
Arega AM, Pattanaik KP, Nayak S, Mahapatra RK (2021) Computational discovery and ex-vivo validation study of novel antigenic vaccine candidates against tuberculosis. Acta Trop 217:105870
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402
Böth D, Steiner EM, Izumi A, Schneider G, Schnell R (2014) RipD (Rv1566c) from Mycobacterium tuberculosis: adaptation of an NlpC/p60 domain to a non-catalytic peptidoglycan-binding function. Biochem J 457(1):33–41
Gustine JN, Au MB, Haserick JR, Hett EC, Rubin EJ, Gibson FC et al (2019) Cell wall hydrolytic enzymes enhance antimicrobial drug activity against mycobacterium. Curr Microbiol 76(4):398–409
Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S et al (2017) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45(D1):D200–D203
Braunstein M, Griffin TJ IV, Kriakov JI, Friedman ST, Grindley NDF, Jacobs WR Jr (2000) Identification of genes encoding exported Mycobacterium tuberculosis proteins using a Tn 552′ phoA in vitro transposition system. J Bacteriol 182(10):2732–2740
Penel S, Arigon AM, Dufayard JF, Sertier AS, Daubin V, Duret L, Gouy M, and Perrière G (2009) June. Databases of homologous gene families for comparative genomics. In BMC bioinformatics (Vol. 10, No. 6, pp. 1–13). BioMed Central
Martinelli DJ, Pavelka MS Jr (2016) The RipA and RipB peptidoglycan endopeptidases are individually nonessential to Mycobacterium smegmatis. J Bacteriol 198(9):1464–1475
Altenhoff AM, Glover NM, Train C-M, Kaleb K, Warwick Vesztrocy A, Dylus D et al (2018) The OMA orthology database in 2018: retrieving evolutionary relationships among all domains of life through richer web and programmatic interfaces. Nucleic Acids Res 46(D1):D477–D485
UniProt Consortium (2019) UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47(D1):D506–D515
Rifaioglu AS, Nalbat E, Atalay V, Martin MJ, Cetin-Atalay R, Doğan T (2020) DEEPScreen: high performance drug–target interaction prediction with convolutional neural networks using 2-D structural compound representations. Chem Sci 11(9):2531–2557
Duffy S, Sykes ML, Jones AJ, Shelper TB, Simpson M, Lang R et al (2017) Screening the Medicines for Malaria Venture Pathogen Box across multiple pathogens reclassifies starting points for open-source drug discovery. Antimicrob Agents Chemother 61(9):e00379-e417
Yang J, Zhang Y (2015) Protein structure and function prediction using I-TASSER. Curr Protoc Bioinforma 52(1):5–8
Schrodinger LLC (2016) The PyMOL molecular graphics system, version 1.3 r1. PyMol.582
Xu D, Zhang Y (2011) Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys J 101(10):2525–2534
Ramachandran GNT, Sasisekharan V (1968) Conformation of polypeptides and proteins. Adv Protein Chem 23:283–437
Laskowski RA, MacArthur MW, Thornton JM (2006) PROCHECK: validation of protein-structure coordinates. International Tables of Crystallography F:722–725
Eisenberg D, Lüthy R, Bowie JU (1997) VERIFY3D: assessment of protein models with three-dimensional profiles. In: Methods in enzymology, vol. 277. Academic Press, pp 396–404
Pontius J, Richelle J, Wodak SJ (1996) Deviations from standard atomic volumes as a quality measure for protein crystal structures. J Mol Biol 264(1):121–136
Colovos C, Yeates TO (1993) ERRAT: an empirical atom-based method for validating protein structures. Protein Sci 2(9):1511–1519
Wass MN, Kelley LA, Sternberg MJE (2010) 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Res. 38(suppl_2):W469-73
Kim S, Chen J, Cheng T, Gindulyte A, He J, He S et al (2019) PubChem 2019 update: improved access to chemical data. Nucleic Acids Res 47(D1):D1102–D1109
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) Open Babel: An open chemical toolbox. J Cheminform 3(1):33
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT et al (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 47(7):1739–49
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS et al (2009) AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem 30(16):2785–2791
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461
Berendsen HJC, van der Spoel D, van Drunen R (1995) GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun 91(1–3):43–56
Da Silva AWS, Vranken WF (2012) ACPYPE-Antechamber python parser interface. BMC Res Notes 5(1):1–8
Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins Struct Funct Bioinforma 65(3):712–725
Hess B, Bekker H, Berendsen HJC, Fraaije JGEM (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18(12):1463–1472
Lemkul JA, Allen WJ, Bevan DR (2010) Practical considerations for building GROMOS-compatible small-molecule topologies. J Chem Inf Model 50(12):2221–2235
Vaught A (1996) Graphing with Gnuplot and Xmgr: two graphing packages available under linux. Linux J. 1996(28es):7
Miller BR III, McGee TD Jr, Swails JM, Homeyer N, Gohlke H, Roitberg AE (2012) MMPBSA. py: an efficient program for end-state free energy calculations. J Chem Theory Comput. 8(9):3314–21
Ertl P (2010) Molecular structure input on the web. J Cheminform 2(1):1–9
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7(1):1–13
Gold B, Roberts J, Ling Y, Quezada LL, Glasheen J, Ballinger E et al (2015) Rapid, semiquantitative assay to discriminate among compounds with activity against replicating or nonreplicating Mycobacterium tuberculosis. Antimicrob Agents Chemother 59(10):6521–6538
Woods GL, Brown-Elliott BA, Conville PS, Desmond EP, Hall GS, Lin G, Pfyffer GE, Ridderhof JC, Siddiqi SH, Wallace Jr RJ, Warren NG (2011) Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes. In: Approved standard—2nd ed, vol 31. Clinical and Laboratory Standards Institute, Wayne
Organization WH (2018) Technical manual for drug susceptibility testing of medicines used in the treatment of tuberculosis. Licence CC BY-NC-SA. 2019;3
Swift ML (1997) GraphPad prism, data analysis, and scientific graphing. J Chem Inf Comput Sci 37(2):411–412
Kapopoulou A, Lew JM, Cole ST (2011) The MycoBrowser portal: a comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis 91(1):8–13
Song H, Sandie R, Wang Y, Andrade-Navarro MA, Niederweis M (2008) Identification of outer membrane proteins of Mycobacterium tuberculosis. Tuberculosis 88(6):526–544
Li J, Cao R, Cheng J (2015) A large-scale conformation sampling and evaluation server for protein tertiary structure prediction and its assessment in CASP11. BMC Bioinformatics 16(1):1–11
Geourjon C, Deleage G (1995) SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 11(6):681–684
Ballell L, Bates RH, Young RJ, Alvarez-Gomez D, Alvarez-Ruiz E, Barroso V et al (2013) Fueling open-source drug discovery: 177 small-molecule leads against tuberculosis. ChemMedChem 8(2):313
Wang E, Sun H, Wang J, Wang Z, Liu H, Zhang JZH et al (2019) End-point binding free energy calculation with MM/PBSA and MM/GBSA: strategies and applications in drug design. Chem Rev 119(16):9478–9508
Kumari R, Kumar R, Consortium OSDD, Lynn A (2014) g_mmpbsa-A GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model 54(7):1951–1962
Horoiwa S, Yokoi T, Masumoto S, Minami S, Ishizuka C, Kishikawa H et al (2019) Structure-based virtual screening for insect ecdysone receptor ligands using MM/PBSA. Bioorg Med Chem 27(6):1065–1075
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 23(1–3):3–25
Kujawski J, Popielarska H, Myka A, Drabińska B, Bernard MK (2012) The log P parameter as a molecular descriptor in the computer-aided drug design–an overview. Comput Methods Sci Technol 18(2):81–88
Veber DF, Johnson SR, Cheng H-Y, Smith BR, Ward KW, Kopple KD (2002) Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 45(12):2615–2623
Baranowski C, Welsh MA, Sham L-T, Eskandarian HA, Lim HC, Kieser KJ et al (2018) Maturing Mycobacterium smegmatis peptidoglycan requires non-canonical crosslinks to maintain shape. Elife 7:e37516
Yuan T, Sampson NS (2018) Hit generation in TB drug discovery: from genome to granuloma. Chem Rev 118(4):1887–1916
Lelovic N, Mitachi K, Yang J, Lemieux MR, Ji Y, Kurosu M (2020) Application of Mycobacterium smegmatis as a surrogate to evaluate drug leads against Mycobacterium tuberculosis. J Antibiot (Tokyo) 73(11):780–789
Andries K, Verhasselt P, Guillemont J, Göhlmann HWH, Neefs J-M, Winkler H et al (2005) A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science (80-) 307(5707):223–7
Dalberto PF, De Souza EV, Abbadi BL, Neves CE, Rambo RS, Ramos AS et al (2020) The many hurdles on the way to anti-tuberculosis drug development. Front Chem 8:984
Yajko DM, Sanders CA, Madej JJ, Cawthon VL, Hadley WK (1996) In vitro activities of rifabutin, azithromycin, ciprofloxacin, clarithromycin, clofazimine, ethambutol, and amikacin in combinations of two, three, and four drugs against Mycobacterium avium. Antimicrob Agents Chemother 40(3):743–749
Acknowledgements
The authors would like to acknowledge Bioinformatics and Immunology laboratory facilities of School of Biotechnology, KIIT Deemed to be University, Bhubaneswar, for completion of the research work. The help from Mr. Somdeb Chatterjee, NII, New Delhi, is also of great importance, particularly during the MD simulation analysis. In addition, the author would like to acknowledge KIIT Deemed to be University, Odisha, India, and Ethiopia Federal Democratic Republic, National Veterinary Institute, Debre Zeit, Ethiopia, for providing the financial support to conduct the PhD research work.
Funding
This research work is not supported by any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
AM contributed to performing and analyzing the in silico and wet-lab experiments as well as writing and revision of the manuscript. AKD performed the MD simulation and analysis of the results. SN provided the chemicals, Petri-plates, and co-supervised the study. RKM conceived the project, supervised the whole study, and corrected the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that no conflict of interest exists.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Arega, A.M., Dhal, A.K., Nayak, S. et al. In silico and in vitro study of Mycobacterium tuberculosis H37Rv uncharacterized protein (RipD): an insight on tuberculosis therapeutics. J Mol Model 28, 171 (2022). https://doi.org/10.1007/s00894-022-05148-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-022-05148-1