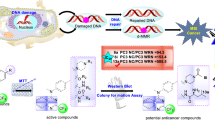
Abstract
Cisplatin is a widely used anti-cancer drug which inhibits the replication and polymerization of DNA molecule while showing some side effects and drug resistance. For this reason, to enhance its therapeutic index, researchers have synthesized several thousand analogs and tested their properties. In this project, several cisplatin analogs were designed to theoretically study the biological activity and lipophilicity effects on amine changes. The amines of the cisplatin molecule were substituted with aliphatic amines in different analogs. Computational methods such as molecular dynamics simulation, molecular docking, and molecular mechanics Poisson-Boltzmann surface area analysis were performed to investigate the binding of six cisplatin derivatives with DNA. The binding affinity and potential interactions of these drugs with double-strand DNA were analyzed. The stability effect of these drugs was investigated via root-mean-square deviation and root-mean-square fluctuation analysis, which showed that some analogs can break base-pair interaction at the end of DNA and reduced the stability of DNA. Also, the results revealed that the hydrogen bond is one of the most important factors in the binding of cisplatin’s adduct to DNA. Molecular mechanics Poisson-Boltzmann surface area analysis indicated that electrostatic and van der Waals interactions are the most important deriving forces to the binding of cisplatin’s drug to DNA. Finally, data revealed that cisplatin and the cis-dichloro-dimethylamine-platin tendency for binding to DNA are greater than that of other analogs.
Graphical abstract








Similar content being viewed by others
Data availability
The datasets generated during and analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.
Code availability
GROMACS software version 5.1.2.
Abbreviations
- MMPBSA:
-
Molecular mechanics Poisson-Boltzmann surface area
- MD simulation:
-
Molecular dynamics simulation
- RMSD:
-
Root-mean-square deviation
- RMSF:
-
Root-mean-square fluctuation
- VdW:
-
Van der Waals
- DNA:
-
Deoxyribonucleic acid
- cis-[Pt(methyl-NH2)2Cl2]:
-
cis-Dichloro-dimethylamine-platin (II)
- cis-[Pt(propyl-NH2)2Cl2]:
-
cis-Dichloro-dipropylamine-platin (II)
- cis-[Pt(butyl-NH2)2Cl2]:
-
cis-Dichloro-dibutylamine-platin (II)
- cis-[Pt(isobutyl-NH2)2Cl2]:
-
cis-Dichloro-diisobutylamine-platin (II)
- cis-[Pt(tertbutyl-NH2)2Cl2]:
-
cis-Dichloro-ditertbutylamine-platin (II)
- cis-[Pt(isopentyl-NH2)2Cl2]:
-
cis-Dichloro-diisopentylamine-platin (II
References
Hambley TW (2001) Platinum binding to DNA: structural controls and consequences. J Chem Soc Dalton Trans 19:2711–2718
Eriksson M, Hassan S, Larsson R, Linder S, Ramqvist T, Lövborg H, Vikinge T, Figgemeier E, Müller J, Stetefeld J, Dalianis T, Ozbek S (2009) Utilization of a right-handed coiled-coil protein from archaebacterium staphylothermus marinus as a carrier for cisplatin. Anticancer Res 29:11–18
Reedijk J (2003) New clues for platinum antitumor chemistry: kinetically controlled metal binding to DNA. Proc Natl Acad Sci 100(7):3611–3616
Michalke B (2010) Platinum speciation used for elucidating activation or inhibition of Pt-containing anti-cancer drugs. J Trace Elem Med Biol 24(2):69–77
Trimmer EE, Essigmann JM (1999) Cisplatin. Essays Biochem 34:191–211
Kelland L (2007) The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer 7(8):573–584
Jamieson ER, Lippard SJ (1999) Structure, recognition, and processing of cisplatin− DNA adducts. Chem Rev 99(9):2467–2498
Chen B, Zhou L (2015) Computational study on mechanisms of the anticancer drug: cisplatin and novel polynuclear platinum (II) interaction with sulfur-donor biomolecules and DNA purine bases. Comput Theor Chem 1074:36–49
Yang D et al (1995) Structure and isomerization of an intrastrand cisplatin-cross-linked octamer DNA duplex by NMR analysis. Biochemistry 34(39):12912–12920
Ciccarelli RB et al (1985) In vivo effects of cis-and trans-diamminedichloroplatinum (II) on SV40 chromosomes: differential repair, DNA-protein crosslinking, and inhibition of replication. Biochemistry 24(26):7533–7540
Jordan P, Carmo-Fonseca M (2000) Molecular mechanisms involved in cisplatin cytotoxicity. Cell Mol Life Sci 57(8–9):1229–1235
Siddik ZH (2003) Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene 22(47):7265–7279
Sherman SE et al (1988) Crystal and molecular structure of cis-[Pt (NH3) 2 [d (pGpG)]], the principal adduct formed by cis-diamminedichloroplatinum (II) with DNA. J Am Chem Soc 110(22):7368–7381
Malinge J-M, Schwartz A, Leng M (1987) Characterization of the ternary complexes formed in the reaction of cis-diamminedichloro platinum (II), ethidium bromide, and nucleic acids. Nucleic Acids Res 15(4):1779–1797
Eastman A (1990) Activation of programmed cell death by anticancer agents: cisplatin as a model system. Cancer Cell 2(8–9):275–280
Fichtinger-Schepman AMJ et al (1985) Adducts of the antitumor drug cis-diamminedichloroplatinum (II) with DNA: formation, identification, and quantitation. Biochemistry 24(3):707–713
Inagaki K, Kidani Y (1983) Studies on the reaction products of guanylyl (3′-5′) adenosine with cis-Pt (NH2)2Cl2and [Pt(NH3)3Cl]Cl. Inorganica Chim Acta 80:171–176
Van Hemelryck B et al (1987) Sequence-dependent platinum chelation by adenylyl (3′-5′) guanosine and guanylyl (3′-5′) adenosine reacting with cis-diamminedichloroplatinum (II) and its diaqua derivative. Inorg Chem 26(6):787–795
Mantri Y, Lippard SJ, Baik M-H (2007) Bifunctional binding of cisplatin to DNA: why does cisplatin form 1, 2-intrastrand cross-links with AG but not with GA? J Am Chem Soc 129(16):5023–5030
Kelland LR (2000) Preclinical perspectives on platinum resistance. Drugs 59(4):1–8
Wilson JJ, Lippard SJ (2014) Synthetic methods for the preparation of platinum anticancer complexes. Chem Rev 114(8):4470–4495
Johnstone TC, Suntharalingam K, Lippard SJ (2016) The next generation of platinum drugs: targeted Pt (II) agents, nanoparticle delivery, and Pt (IV) prodrugs. Chem Rev 116(5):3436–3486
Weiss RB, Christian MC (1993) New cisplatin analogues in development. Drugs 46(3):360–377
Shah N, Dizon DS (2009) New-generation platinum agents for solid tumors. Future Oncol 5(1):33–42
Wang D, Lippard SJ (2005) Cellular processing of platinum anticancer drugs. Nat Rev Drug Disc 4(4):307–320
Kelland LR (1993) New platinum antitumor complexes. Crit Rev Oncol Hematol 15(3):91–219
Rixe O et al (1996) Oxaliplatin, tetraplatin, cisplatin, and carboplatin: spectrum of activity in drug-resistant cell lines and in the cell lines of the National Cancer Institute’s Anticancer Drug Screen panel. Biochem Pharmacol 52(12):1855–1865
Alberto ME, Butera V, Russo N (2011) Which one among the Pt-containing anticancer drugs more easily forms monoadducts with G and A DNA bases? A comparative study among oxaliplatin, nedaplatin, and carboplatin. Inorg Chem 50(15):6965–6971
Natarajan G, Malathi R, Holler E (1999) Increased DNA-binding activity of cis-1, 1-cyclobutanedicarboxylatodiammineplatinum (II)(carboplatin) in the presence of nucleophiles and human breast cancer MCF-7 cell cytoplasmic extracts: activation theory revisited. Biochem Pharmacol 58(10):1625–1629
Raymond E, Faivre S, Woynarowski JM, Chaney SG (1998) Oxaliplatin: mechanism of action and antineoplastic activity. Semin Oncol 25(2 Suppl 5):4–12
Wiseman LR et al (1999) Oxaliplatin. Drugs Aging 14(6):459–475
Alcindor T, Beauger N (2011) Oxaliplatin: a review in the era of molecularly targeted therapy. Current Oncol (Toronto, Ont.) 18(1):18–25
Wu Y et al (2007) Solution structures of a DNA dodecamer duplex with and without a cisplatin 1, 2-d (GG) intrastrand cross-link: comparison with the same DNA duplex containing an oxaliplatin 1, 2-d (GG) intrastrand cross-link. Biochemistry 46(22):6477–6487
Spiegel K, Rothlisberger U, Carloni P (2004) Cisplatin binding to DNA oligomers from hybrid Car-Parrinello/molecular dynamics simulations. J Phys Chem B 108(8):2699–2707
Scheeff ED, Briggs JM, Howell SB (1999) Molecular modeling of the intrastrand guanine-guanine DNA adducts produced by cisplatin and oxaliplatin. Mol Pharmacol 56(3):633–643
Magistrato A et al (2006) Binding of novel azole-bridged dinuclear platinum (II) anticancer drugs to DNA: insights from hybrid QM/MM molecular dynamics simulations. J Phys Chem B 110(8):3604–3613
Elizondo-Riojas M-A, Kozelka J (2001) Unrestrained 5 ns molecular dynamics simulation of a cisplatin-DNA 1, 2-GG adduct provides a rationale for the NMR features and reveals increased conformational flexibility at the platinum binding site. J Mol Biol 314(5):1227–1243
Olusanya TO et al (2018) Liposomal drug delivery systems and anticancer drugs. Molecules 23(4):907
Liu D et al (2013) Application of liposomal technologies for delivery of platinum analogs in oncology. Int J Nanomed 8:3309
Zalba S, Garrido MJ (2013) Liposomes, a promising strategy for clinical application of platinum derivatives. Expert Opin Drug Deliv 10(6):829–844
Ghosh S (2019) Cisplatin: the first metal based anticancer drug. Bioorg Chem 88:102925. https://doi.org/10.1016/j.bioorg.2019.102925
Yassin AM, Elnouby M, El-Deeb NM, Hafez EE (2016) Tungsten oxide nanoplates; the novelty in targeting metalloproteinase-7 gene in both cervix and colon cancer cells. Appl Biochem Biotechnol 180(4):623–637. https://doi.org/10.1007/s12010-016-2120-x
Kesharwani RK, Srivastava V, Singh P, Rizvi SI, Adeppa K, Misra K (2015) A novel approach for overcoming drug resistance in breast cancer chemotherapy by targeting new synthetic curcumin analogues against aldehyde dehydrogenase 1 (ALDH1A1) and glycogen synthase kinase-3 β (GSK-3β). Appl Biochem Biotechnol 176(7):1996–2017. https://doi.org/10.1007/s12010-015-1696-x
Hardie ME, Kava HW, Murray V (2016) Cisplatin analogues with an increased interaction with DNA: prospects for therapy. Curr Pharm Des 22(44):6645–6664
Frisch M et al (2016) Gaussian 16 Rev. B. 01, Wallingford, CT
Berman HM et al (2002) The protein data bank. Acta Crystallogr D Biol Crystallogr 58(6):899–907
Morris GM et al (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19(14):1639–1662
Huey R et al (2007) A semiempirical free energy force field with charge-based desolvation. J Comput Chem 28(6):1145–1152
DeLano WL (2002) PyMOL: an open-source molecular graphics tool, CCP4 newsletter pro. Crystallogr 40(1):82–92
Pettersen EF et al (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25(13):1605–1612
Hess B et al (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4(3):435–447
Wang J et al (2004) Development and testing of a general amber force field. J Comput Chem 25(9):1157–1174
Lindorff-Larsen K et al (2010) Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins Struct Funct Bioinf 78(8):1950–1958
Yesylevskyy S et al (2015) Empirical force field for cisplatin based on quantum dynamics data: case study of new parameterization scheme for coordination compounds. J Mol Model 21(10):268
Lopes JF et al (2006) Monte Carlo simulation of cisplatin molecule in aqueous solution. J Phys Chem B 110(24):12047–12054
Paschoal D et al (2012) The role of the basis set and the level of quantum mechanical theory in the prediction of the structure and reactivity of cisplatin. J Comput Chem 33(29):2292–2302
Bussi G, Donadio D, Parrinello M (2007) Canonical sampling through velocity rescaling. J Chem Phys 126:014101
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an N⋅ log (N) method for Ewald sums in large systems. J Chem Phys 98(12):10089–10092
Andersen HC (1983) Rattle: a “velocity” version of the shake algorithm for molecular dynamics calculations. J Comput Phys 52(1):24–34
Miyamoto S, Kollman PA (1992) Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J Comput Chem 13(8):952–962
Kumari R et al (2014) gmmpbsa: a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model 54(7):1951–1962
Funding
Chemistry & Chemical Engineering Research Center of Iran financially supported this project.
Author information
Authors and Affiliations
Contributions
The authors confirm contribution to the paper as follows: study conception and design: Mahboube Eslami Moghadam; data collection, analysis, and interpretation of results: Arezo Rahiminezhad and Mahboube Eslami Moghadam; draft manuscript preparation: Arezo Rahiminezhad, Mahboube Eslami Moghadam, Adeleh Divsalar, and A. Vahid Mesbah. All authors reviewed the results and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rahiminezhad, A., Moghadam, M.E., Divsalar, A. et al. How can the cisplatin analogs with different amine act on DNA during cancer treatment theoretically?. J Mol Model 28, 2 (2022). https://doi.org/10.1007/s00894-021-04984-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-021-04984-x