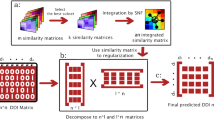
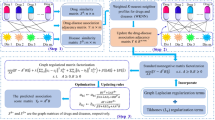
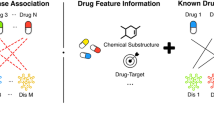
Abstract
Due to rising development costs and stagnant product outputs of traditional drug discovery methods, drug repositioning, which discovers new indications for existing drugs, has attracted increasing interest. Computational drug repositioning can integrate prioritization information and accelerate time lines even further. However, most existing methods for predicting drug repositioning have low precisions. The present article proposed a new method named DDAPRED (https://github.com/nongdaxiaofeng/DDAPRED) for drug repositioning prediction. The method integrated multiple sources of drug similarity and disease similarity information, and it used the regularized logistic matrix decomposition method to significantly improve the prediction performance. In 5-fold cross-validation, the areas under the receiver operating characteristic curve (AUROC) and the precision-recall curve (AUPRC) of DDAPRED reached 0.932 and 0.438, respectively, exceeding other methods. The present study also analyzed the parameters influencing the model performance and the effect of different drug similarity information in-depth, and it verified the treatment relationship of the top 50 predictions with unknown relationships in the training set, further demonstrating the practicability of our method.



Similar content being viewed by others
References
Hurle MR, Yang L, Xie Q, Rajpal DK, Sanseau P, Agarwal P (2013) Computational drug repositioning: from data to therapeutics. Clin Pharmacol Ther 93(4):335–341
Sleigh SH, Barton CL (2010) Repurposing strategies for therapeutics. Pharmaceut Med 24(3):151–159
Ashburn TT, Thor KB (2004) Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov 3:673
Soignet SL, Maslak P, Wang Z-G, Jhanwar S, Calleja E, Dardashti LJ, Corso D, DeBlasio A, Gabrilove J, Scheinberg DA et al (1998) Complete remission after treatment of acute promyelocytic leukemia with arsenic trioxide. New Engl J Med 339(19):1341–1348
Swanson DR (1990) Medical literature as a potential source of new knowledge. Bull Med Libr Assoc 78(1):29–37
Chang A, Butte A (2009) Systematic evaluation of drug–disease relationships to identify leads for novel drug uses. Clin Pharmacol Ther 86(5):507–510
Gottlieb A, Stein GY, Ruppin E, Sharan R (2011) PREDICT: a method for inferring novel drug indications with application to personalized medicine. Mol Syst Biol 7(1):496
Wang Y, Chen S, Deng N, Wang Y (2013) Drug repositioning by kernel-based integration of molecular structure, molecular activity, and phenotype data. PLoS One 8(11):e78518
Wang W, Yang S, Zhang X, Li J (2014) Drug repositioning by integrating target information through a heterogeneous network model. Bioinformatics 30(20):2923–2930
Liang X, Zhang P, Yan L, Fu Y, Peng F, Qu L, Shao M, Chen Y, Chen Z (2017) LRSSL: predict and interpret drug–disease associations based on data integration using sparse subspace learning. Bioinformatics 33(8):1187–1196
Zhang W, Yue X, Lin W, Wu W, Liu R, Huang F, Liu F (2018) Predicting drug-disease associations by using similarity constrained matrix factorization. BMC Bioinformatics 19(1):233
Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K (2017) KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45(D1):D353–D361
Mathur S, Dinakarpandian D (2012) Finding disease similarity based on implicit semantic similarity. J Biomed Inform 45(2):363–371
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT et al (2000) Gene ontology: tool for the unification of biology. Nat Genet 25:25
van Laarhoven T, Nabuurs SB, Marchiori E (2011) Gaussian interaction profile kernels for predicting drug–target interaction. Bioinformatics 27(21):3036–3043
Jaccard P (1912) The distribution of the flora in the alpine zone.1. New Phytol 11(2):37–50
Wang B, Mezlini AM, Demir F, Fiume M, Tu Z, Brudno M, Haibe-Kains B, Goldenberg A (2014) Similarity network fusion for aggregating data types on a genomic scale. Nat Methods 11:333
Ban T, Ohue M, Akiyama Y (2019) NRLMFβ: Beta-distribution-rescored neighborhood regularized logistic matrix factorization for improving the performance of drug–target interaction prediction. Biochem Biophys Rep 18:100615
Liu Y, Wu M, Miao C, Zhao P, Li X-L (2016) Neighborhood regularized logistic matrix factorization for drug-target interaction prediction. PLoS Comp Biol 12(2):e1004760
He B-S, Qu J, Zhao Q (2018) Identifying and exploiting potential miRNA-disease associations with neighborhood regularized logistic matrix factorization. Front Genet 9:303
Wang X, Yan R, Li J, Song J (2016) SOHPRED: a new bioinformatics tool for the characterization and prediction of human S-sulfenylation sites. Mol BioSyst 12(9):2849–2858
Fan J, Upadhye S, Worster A (2015) Understanding receiver operating characteristic (ROC) curves. CJEM 8(1):19–20
Boyd K, Eng KH, Page CD (2013) Area under the precision-recall curve: point estimates and confidence intervals. In: Blockeel H. et al (eds) Machine Learning and Knowledge Discovery in Databases. ECML PKDD 2013. Lecture Notes in Computer Science, pp 451–466
Saito T, Rehmsmeier M (2015) The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS One 10(3):e0118432
Zheng X, Ding H, Mamitsuka H, Zhu S (2013) Collaborative matrix factorization with multiple similarities for predicting drug-target interactions. In: Proceedings of the 19th ACM SIGKDD international conference on Knowledge discovery and data mining. Chicago, Illinois, USA, p 1025–1033. 2487670: ACM
Luo Y, Zhao X, Zhou J, Yang J, Zhang Y, Kuang W, Peng J, Chen L, Zeng J (2017) A network integration approach for drug-target interaction prediction and computational drug repositioning from heterogeneous information. Nat Commun 8(1):573
McMurray JJ (2010) Systolic heart failure. N Engl J Med 362(3):228–238
Richards DM, Heel RC, Brogden RN, Speight TM, Avery GS (1984) Ceftriaxone. Drugs 27(6):469–527
De Cueto M, Sanchez M-J, Sampedro A, Miranda J-A, Herruzo A-J, Rosa-Fraile M (1998) Timing of intrapartum ampicillin and prevention of vertical transmission of group B streptococcus. Obstet Gynecol 91(1):112–114
Chen X, Yan CC, Zhang X, Zhang X, Dai F, Yin J, Zhang Y (2016) Drug-target interaction prediction: databases, web servers and computational models. Brief Bioinform 17(4):696–712
Liu Y, Zeng X, He Z, Zou Q (2017) Inferring microRNA-disease associations by random walk on a heterogeneous network with multiple data sources. IEEE/ACM Trans Comput Biol Bioinform 14(4):905–915
Perozzi B, Al-Rfou R, Skiena S (2014) DeepWalk: online learning of social representations. In: Proceedings of the 20th ACM SIGKDD international conference on Knowledge discovery and data mining. New York, New York, USA, p 701–710. 2623732: ACM
Grover A, Leskovec J (2016) node2vec: scalable feature learning for networks. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. San Francisco, California, USA, p 855–864. 2939754: ACM
Schlichtkrull M, Kipf TN, Bloem P, Berg R, Titov I, Welling M (2018) Modeling relational data with graph convolutional networks. In: Gangemi A. et al. (eds) The Semantic Web. ESWC 2018. Lecture Notes in Computer Science, pp 593–607
Funding
This work was supported by the National Natural Science Foundation of China (31500673 and 31571300).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 27 kb).
Rights and permissions
About this article
Cite this article
Wang, X., Yan, R. DDAPRED: a computational method for predicting drug repositioning using regularized logistic matrix factorization. J Mol Model 26, 60 (2020). https://doi.org/10.1007/s00894-020-4315-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-020-4315-x