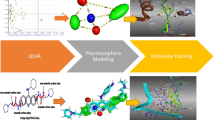
Abstract
Rising mortality due to cancer has led to the development and identification of newer targets and molecules to cure the disease. Telomerase is one of the attractive targets for design of many chemotherapeutic drugs. This research highlights the designing of novel telomerase inhibitors using ligand-based (3D-QSAR) and structure-based (molecular docking and molecular dynamics simulation) approaches. For the development of the 3D-QSAR model, 37 synthetic molecules reported earlier as telomerase inhibitors were selected from diversified literature. Three different alignment methods were explored; among them, distill alignment was found to be the best method with good statistical results and was used for the generation of QSAR model. Statistically significant CoMSIA model with a correlation coefficient (r2ncv) value of 0.974, leave one out (q2) value of 0.662 and predicted correlation coefficient (r2pred) value of 0.560 was used for the analysis of QSAR. For the MDS study, A-chain of telomerase was stabilised for 50 ns with respect to 1-atm pressure, with an average temperature of 299.98 k and with potential energy of 1,145,336 kJ/m converged in 997 steps. Furthermore, the behaviour study of variants towards the target revealed that active variable gave better affinity without affecting amino acid sequences and dimensions of protein which was accomplished through RMSD, RMSF and Rg analysis. Results of molecular docking study supported the outcomes of QSAR contour maps as ligand showed similar interactions with surrounded amino acids which were identified in contour map analysis. The results of the comprehensive study might be proved valuable for the development of potent telomerase inhibitors.
Graphical abstract












Similar content being viewed by others
References
Puri N, Girard J (2013) Novel therapeutics targeting telomerase and telomeres. J Cancer Sci Ther 5:1–3. https://doi.org/10.4172/1948-5956.1000e127
Tang H, Wang H et al (2018) HuR regulates telomerase activity through TERC methylation. Nat Commun 9:1–12. https://doi.org/10.1038/s41467-018-05213-5
Ganesan K, Xu B (2018) Telomerase inhibitors from natural products and their anticancer potential. Int J Mol Sci 19. https://doi.org/10.3390/ijms19010013
Jäger K, Walter M (2016) Therapeutic targeting of telomerase, Genes (Basel). 7 1–24. https://doi.org/10.3390/genes7070039
Agrawal A, Dang S, Gabrani R (2012) Recent patents on anti-telomerase cancer therapy, Recent Pat. Anti-cancer. Drug Discov 7:102–117. https://doi.org/10.2174/157489212798357958
Zhang L, Huang J, Ren L et al (2008) Synthesis and evaluation of cationic phthalocyanine derivatives as potential inhibitors of telomerase. Bioorg Med Chem 16:303–312. https://doi.org/10.1016/j.bmc.2007.09.037
Wang J, Liu L, Ma H (2017) Sensors and actuators B: chemical label-free real-time investigation of the effect of telomerase inhibitors based on quartz crystal microbalance measurement. Sensors Actuators B Chem 239:943–950. https://doi.org/10.1016/j.snb.2016.08.021
Seimiya H, Oh-Hara T, Suzuki T et al (2002) Telomere shortening and growth inhibition of human cancer cells by novel synthetic telomerase inhibitors MST-312, MST-295, and MST-199 1. Mol Cancer Ther 1:657–665 http://mct.aacrjournals.org/content/molcanther/1/9/657.full.pdf. Accessed 29 Dec 2017
Drewe WC, Nanjunda R, Gunaratnam M et al (2008) Rational design of substituted diarylureas: a scaffold for binding to G-quadruplex motifs. J Med Chem 51:7751–7767. https://doi.org/10.1021/jm801245v
Wang Y, Cheng FX, Yuan XL et al (2016) Dihydropyrazole derivatives as telomerase inhibitors: structure-based design, synthesis, SAR and anticancer evaluation in vitro and in vivo. Eur J Med Chem 112:231–251. https://doi.org/10.1016/J.EJMECH.2016.02.009
Verma J, Khedkar VM, Coutinho EC (2010) 3D-QSAR in drug design-a review. Curr Top Med Chem 10:95–115. https://doi.org/10.2174/156802610790232260
Zambre VP, Murumkar PR, Giridhar R, Yadav MR (2010) Development of highly predictive 3D-QSAR CoMSIA models for anthraquinone and acridone derivatives as telomerase inhibitors targeting G-quadruplex DNA telomere. J Mol Graph Model 29:229–239. https://doi.org/10.1016/j.jmgm.2010.07.003
Halim SA, Ul-Haq Z (2015) Structure based 3D-QSAR studies of Interleukin-2 inhibitors: comparing the quality and predictivity of 3D-QSAR models obtained from different alignment methods and charge calculations. Chem Biol Interact 238:9–24. https://doi.org/10.1016/j.cbi.2015.05.018
Feng K, Ren Y, Li R (2017) Combined pharmacophore-guided 3D-QSAR, molecular docking and molecular dynamics studies for evodiamine analogs as DNA topoisomerase I inhibitors, J. Taiwan Inst. Chem Eng 78:81–95. https://doi.org/10.1016/j.jtice.2017.06.027
Patel B. D, Ghate M. D, (2015) 3D-QSAR studies of dipeptidyl peptidase-4 inhibitors using various alignment methods, Med Chem Res 241060–1069. https://doi.org/10.1007/s00044-014-1178-7.
Ismail S, Mohamed AO, Abdel F et al (2012) CoMFA and CoMSIA Studies of 1,2-dihydropyridine derivatives as anticancer agents. Med Chem (Los Angeles) 8(2012):372–383. https://doi.org/10.1097/COC.0b013e3182a79009.Pain
Balupuri A, Balasubramanian PK, Cho SJ (2017) 3D-QSAR, docking, molecular dynamics simulation and free energy calculation studies of some pyrimidine derivatives as novel JAK3 inhibitors. Arab J Chem 09:1–20. https://doi.org/10.1016/j.arabjc.2017.09.009
Alamri MA (2020) Pharmacoinformatics and molecular dynamic simulation studies to identify potential small-molecule inhibitors of WNK-SPAK/OSR1 signaling that mimic the RFQV motifs of WNK kinases. Arab J Chem 13:5107–5117. https://doi.org/10.1016/j.arabjc.2020.02.010
Sargsyan K, Grauffel C, Lim C (2017) How molecular size impacts RMSD applications in molecular dynamics simulations. J Chem Theory Comput 13:1518–1524. https://doi.org/10.1021/acs.jctc.7b00028
Ho BK, Brasseur R (2005) The Ramachandran plots of glycine and pre-proline. BMC Struct Biol 5:1–11. https://doi.org/10.1186/1472-6807-5-14
Fan Z, Ho S, Wen R et al (2019) Design, synthesis and molecular docking analysis of flavonoid derivatives as potential. Molecules. 24:1–14. https://doi.org/10.3390/molecules24173180
Pereira GRC, Tavares GDB, Freitas MC, Mesquita JF (2020) In silico analysis of the tryptophan hydroxylase 2 (TPH2) protein variants related to psychiatric disorders. PLoS One 15:1–23. https://doi.org/10.1371/journal.pone.0229730
Pagadala NS, Syed K, Tuszynski J (2017) Software for molecular docking: a review. Biophys Rev 9:91–102. https://doi.org/10.1007/s12551-016-0247-1
Duan Y, Yao Y, Tang D (2014) Synthesis and biological evaluation of quinoline–imidazole hybrids as potent telomerase inhibitors: a promising class of antitumor agents, R Soc Chem 420382–20392. https://doi.org/10.1039/c4ra01936a
Zhou J, Lu Y, Ou T, Zhou J, Huang Z, Zhu X, Du C (2005) Synthesis and evaluation of quindoline derivatives as g-quadruplex inducing and stabilizing ligands and potential inhibitors of telomerase. J Med Chem 48:7315–7321. https://doi.org/10.1021/jm050041b
Sun J, Zhu H, Yang Z, Zhu H (2013) Synthesis, molecular modeling and biological evaluation of 2-aminomethyl-5-anticancer agent, Eur J Med Chem 6023–28. https://doi.org/10.1016/j.ejmech.2012.11.039
Harrison RJ, Gowan SM, Kelland LR, Neidle S (1999) Human telomerase inhibition by substituted acridine derivatives, Bioorganic Med. Chem Lett 9:2463–2468. https://doi.org/10.1016/S0960-894X(99)00394-7
Borisa A, Bhatt H (2015) 3D-QSAR (CoMFA, CoMFA-RG, CoMSIA) and molecular docking study of thienopyrimidine and thienopyridine derivatives to explore structural requirements for aurora-B kinase inhibition. Eur J Pharm Sci 79:1–12. https://doi.org/10.1016/j.ejps.2015.08.017
Yadav DK, Saloni, Sharma P et al (2017) Studies of the benzopyran class of selective COX-2 inhibitors using 3D-QSAR and molecular docking. Arch Pharm Res 1–12. https://doi.org/10.1007/s12272-017-0945-7
Chekkara R, Kandakatla N, Gorla VR et al (2017) Theoretical studies on benzimidazole and imidazo[1,2-a] pyridine derivatives as Polo-like kinase 1 (Plk1) inhibitors: pharmacophore modeling, atom-based 3D-QSAR and molecular docking approach. J Saudi Chem Soc 21:S311–S321. https://doi.org/10.1016/j.jscs.2014.03.007
Bryan C, Rice C, Harkisheimer M et al (2015) Structural basis of telomerase inhibition by the article structural basis of telomerase inhibition by the highly specific BIBR1532. Struct Des 23:1934–1942. https://doi.org/10.1016/j.str.2015.08.006
Díaz A, Martínez E, Puerta L et al (2014) A CoMSIA study to design antagonist ligands for the LuxS protein, New J. Chem. 38:1235–1249. https://doi.org/10.1039/c3nj01162c
Chaube U, Chhatbar D, Bhatt H (2016) 3D-QSAR, molecular dynamics simulations and molecular docking studies of benzoxazepine moiety as mTOR inhibitor for the treatment of lung cancer. Bioorg Med Chem Lett 26:864–874. https://doi.org/10.1016/j.bmcl.2015.12.075
Gao J, Sun J, Wang T, Sheng S, Huang T (2017) Combined 3D-QSAR modeling and molecular docking study on spiro-derivatives as inhibitors of acetyl-CoA carboxylase. Med Chem Res 26:361–371. https://doi.org/10.1007/s00044-016-1743-3
Duan YT, Yao YF, Tang DJ (2014) Synthesis and biological evaluation of quinoline-imidazole hybrids as potent telomerase inhibitors: a promising class of antitumor agents. Rsc Adv 4:20382–20392. https://doi.org/10.1039/c4ra01936a
Wang JL, Cheng LP, Wang TC, Deng W, Wu FH (2017) Molecular modeling study of CP-690550 derivatives as JAK3 kinase inhibitors through combined 3D-QSAR, molecular docking, and dynamics simulation techniques. J Mol Graph Model 72:178–186. https://doi.org/10.1016/j.jmgm.2016.12.020
Zhu J, Ke K, Xu L, Jin J (2019) Theoretical studies on the selectivity mechanisms of PI3Kδ inhibition with marketed idelalisib and its derivatives by 3D-QSAR, molecular docking, and molecular dynamics simulation. J Mol Model 25. https://doi.org/10.1007/s00894-019-4129-x
Murumkar P, Sharma MK, Miniyar P. B, Yadav M. R (2016) Development of a credible 3D-QSAR CoMSIA model and docking studies for a series of triazoles and tetrazoles containing 11 β-HSD1 inhibitors, SAR QSAR Environ Res 1–28. https://doi.org/10.1080/1062936X.2016.1167774
Shirgahi TF, Bagherzadeh K, Golestanian S, Jarstfer M (2015) Potent Human telomerase inhibitors: molecular dynamic simulations, multiple pharmacophore-based virtual screening, and biochemical assays. J Chem Inf Model 55:2596–2610. https://doi.org/10.1021/acs.jcim.5b00336
Fuggetta MP, De Mico A, Cottarelli A et al (2016) Synthesis and enantiomeric separation of a novel spiroketal derivative: a potent human telomerase inhibitor with high in vitro anticancer activity. J Med Chem 1–49. https://doi.org/10.1021/acs.jmedchem.6b01046
Acknowledgements
Authors are thankful to Nirma University, Ahmedabad, India, for supporting work, which is a part of Doctor of Philosophy (PhD) research work of Keerti Vishwakarma, to be submitted to Nirma University, Ahmedabad, India.
Author information
Authors and Affiliations
Contributions
Keerti Vishwakarma: conceptualization; methodology and validation; data curation; writing—original draft
Hardik Bhatt: conceptualization; reviewing the methodology, results, and discussion; editing—final draft
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1.
Supplementary Information file contains Field contributors of CoMSIA; Ramachandran plot of telomerase: (a) Plot of unsimulated protein structure; (b) Plot of simulated protein structure; Docking Score of potent molecules along with design compounds by Surflex docking module in SYBYL. (DOCX 853 kb)
ESM 2.
Mol 2 structures are also provided in zip file. (RAR 61.2 kb)
Rights and permissions
About this article
Cite this article
Vishwakarma, K., Bhatt, H. Molecular modelling of quinoline derivatives as telomerase inhibitors through 3D-QSAR, molecular dynamics simulation, and molecular docking techniques. J Mol Model 27, 30 (2021). https://doi.org/10.1007/s00894-020-04648-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-020-04648-2