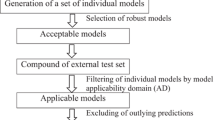
Abstract
Many chemical phenomena occur in solution. Different solvents can change the optical activity of chiral molecules. The optical rotation angles of solutes of 75 amino acids in dimethylformamide, water and methanol were analyzed using the quantitative structure–activity relationships approach. For an accurate description of chirality, we used specific quantum chemical descriptors, which reflect the properties of a chiral center, and continuous symmetry measures. The set of specific quantum chemical descriptors for atoms located near the chiral center, such as Mulliken charges, the sum of Mulliken charges on an atom (with the hydrogen charges summed up with the adjacent non-hydrogen atoms), and nuclear magnetic resoncance tensors was applied. To represent solvent effects, we used mixture-like structural simplex descriptors and quantum chemical descriptors obtained for structures optimized for specified solvent using PBE1PBE/6-31G** level of theory with the polarizable continuum model. Multiple linear regression, M5P, and locally weighted learning techniques were used to achieve accurate predictions. The specific quantum chemical descriptors proposed here demonstrated high specificity in the majority of the developed models and established direct quantitative structure–property relationships.






Similar content being viewed by others
References
Norden B (1977) Was photoresolution of amino acids the origin of optical activity in life? Nature 266:567–568
Randić M, Razinger M (1996) Molecular shapes and chirality. J Chem Inf Comput Sci 36:429–441. https://doi.org/10.1021/ci950091x
de Julián-Ortiz JV, de Gregorio Alapont C, Rios-Santamarina I et al (1998) Prediction of properties of chiral compounds by molecular topology. J Mol Graph Model 16:14–18. https://doi.org/10.1016/S1093-3263(98)00013-8
Vol’kenshtein MV, Kruchek MP (1961) Optical activity of amino acids. J Struct Chem 2:49–52. https://doi.org/10.1007/BF00744855
Zemlicka J (2000) Enantioselectivity of the antiviral effects of nucleoside analogues. Pharmacol Ther 85:251–266
Maury G (2000) The enantioselectivity of enzymes involved in current antiviral therapy using nucleoside analogues: a new strategy? Antivir Chem Chemother 165–189
Mathé C, Gosselin G (2006)l-Nucleoside enantiomers as antivirals drugs: a mini-review. Antiviral Res 71:276–281. https://doi.org/10.1016/j.antiviral.2006.04.017
Dearden JC (2016) The history and development of quantitative structure-activity relationships (QSARs). Oncol Break Res Pract Break Res Pract 67. https://doi.org/10.4018/IJQSPR.2016010101
Gramatica P, Sangion A (2016) A historical excursus on the statistical validation parameters for QSAR models: a clarification concerning metrics and terminology. J Chem Inf Model 56:1127–1131
Cherkasov A, Muratov EN, Fourches D, Varnek A, Baskin II, Cronin M, Dearden J, Gramatica P, Martin YC, Todeschini R, Consonni V, Kuz'min VE, Cramer R, Benigni R, Yang C, Rathman J, Terfloth L, Asteiger J, Richard A, Tropsha A (2014) QSAR modeling: where have you been? Where are you going to? J Med Chem 57(12):4977–5010. https://doi.org/10.1021/jm4004285
Pogliani L (1994) Structure property relationships of amino acids and some dipeptides. Amino Acids 6:141–153
Pogliani L (1993) Molecular connectivity model for determination of physicochemical properties of. alpha.-amino acids. J Phys Chem 97:6731–6736
Liu HX, Zhang RS, Yao XJ et al (2004) Prediction of the isoelectric point of an amino acid based on GA-PLS and SVMs. J Chem Inf Comput Sci 44:161–167
Tham SY, Agatonovic-Kustrin S (2002) Application of the artificial neural network in quantitative structure–gradient elution retention relationship of phenylthiocarbamyl amino acids derivatives. J Pharm Biomed Anal 28:581–590
Zaliani A, Gancia E (1999) MS-WHIM scores for amino acids: a new 3D-description for peptide QSAR and QSPR studies. J Chem Inf Comput Sci 39:525–533
Lin Z, Long H, Bo Z et al (2008) New descriptors of amino acids and their application to peptide QSAR study. Peptides 29:1798–1805
Tian F, Zhou P, Li Z (2007) T-scale as a novel vector of topological descriptors for amino acids and its application in QSARs of peptides. J Mol Struct 830:106–115
Tong J, Liu S, Zhou P et al (2008) A novel descriptor of amino acids and its application in peptide QSAR. J Theor Biol 253:90–97
Fauchère J-L, Lauterwein J (1985) The chemical shift of the alpha carbon in amino-acids as a parameter for QSAR studies of oligopeptides. Quant Struct Relationships 4:11–13. https://doi.org/10.1002/qsar.19850040103
Mennucci B, Tomasi J, Cammi R et al (2002) Polarizable Continuum Model (PCM) calculations of solvent effects on optical rotations of chiral molecules. J Phys Chem A 106:6102–6113. https://doi.org/10.1021/jp020124t
Mukhopadhyay P, Zuber G, Goldsmith M-R et al (2006) Solvent effect on optical rotation: a case study of methyloxirane in water. ChemPhysChem 7:2483–2486. https://doi.org/10.1002/cphc.200600477
Eliel EL, Wilen SH (1994) Stereochemistry of organic compounds. Wiley, New York
Sarmah P, Deka RC (2009) DFT-based QSAR and QSPR models of several cis-platinum complexes: solvent effect. J Comput Aided Mol Des 23:343–354. https://doi.org/10.1007/s10822-009-9265-4
Karelson M, Lobanov VS, Katritzky AR (1996) Quantum-chemical descriptors in QSAR/QSPR studies. Chem Rev 96:1027–1044. https://doi.org/10.1021/cr950202r
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenb DJ (2009) Gaussian 09, Gaussian, Inc., Wallingford CT
Miertuš S, Scrocco E, Tomasi J (1981) Electrostatic interaction of a solute with a continuum. A direct utilizaion of AB initio molecular potentials for the prevision of solvent effects. Chem Phys 55:117–129. https://doi.org/10.1016/0301-0104(81)85090-2
Okovytyy S, Kopteva S, Voronkov E et al (2013) 1H NMR spectra of N-Methyl-4-Tolyl-1-(4-Bromonaphthyl)-Amine and N-Phenyl-1-(4-Bromonaphthyl)-Amine: a combined experimental and theoretical study. Bull Dnipropetr Univ Chem 21:7–15
Verma RP, Hansch C (2011) Use of 13C NMR chemical shift as QSAR/QSPR descriptor. Chem Rev 111:2865–2899. https://doi.org/10.1021/cr100125d
Zabrodsky H, Peleg S, Avnir D (1992) Continuous symmetry measures. J Am Chem Soc 114:7843–7851. https://doi.org/10.1021/ja00046a033
Kuz’min V, Artemenko A, Muratov E (2008) Hierarchical QSAR technology based on the Simplex representation of molecular structure. J Comput Aided Mol De. 22:403–421
Muratov EN, Varlamova EV, Artemenko AG et al (2012) Existing and developing approaches for QSAR analysis of mixtures. Mol Inform 31:202–221. https://doi.org/10.1002/minf.201100129
KNIME.com (2017) KNIME https://www.knime.com/
Gramatica P, Chirico N, Papa E et al (2013) QSARINS: a new software for the development, analysis, and validation of QSAR MLR models. J Comput Chem 34:2121–2132. https://doi.org/10.1002/jcc.23361
Frank E, Hall M, Pfahringer B (2003) Locally weighted naive bayes. In: Proceedings of the Conference on Uncertainty in Artificial Intelligence. Kaufmann, San Francisco, pp 249–256
Jones DE, Ghandehari H, Facelli JC (2016) A review of the applications of data mining and machine learning for the prediction of biomedical properties of nanoparticles. Comput Methods Programs Biomed 132:93–103. https://doi.org/10.1016/j.cmpb.2016.04.025
Chirico N, Gramatica P (2011) Real external predictivity of QSAR models: how to evaluate it? Comparison of different validation criteria and proposal of using the concordance correlation coefficient. J Chem Inf Model 51:2320–2335. https://doi.org/10.1021/ci200211n
Gadaleta D, Mangiatordi GF, Catto M et al (2016) Applicability domain for QSAR models: where theory meets reality. Int J Quant Struct Relationships 1:45–63
Roy K, Ambure P, Aher RB (2017) How important is to detect systematic error in predictions and understand statistical applicability domain of QSAR models? Chemom Intell Lab Syst 162:44–54. https://doi.org/10.1016/j.chemolab.2017.01.010
Aniceto N, Freitas AA, Bender A, Ghafourian T (2016) A novel applicability domain technique for mapping predictive reliability across the chemical space of a QSAR: reliability-density neighbourhood. J Cheminform 8:69. https://doi.org/10.1186/s13321-016-0182-y
Cao D-S, Deng Z-K, Zhu M-F et al (2017) Ensemble partial least squares regression for descriptor selection, outlier detection, applicability domain assessment, and ensemble modeling in QSAR/QSPR modeling. J Chemom 31:e2922. https://doi.org/10.1002/cem.2922
Miller J, Parker AJ (1961) Dipolar aprotic solvents in bimolecular aromatic nucleophilic substitution reactions. J Am Chem Soc 83:117–123. https://doi.org/10.1021/ja01462a023
Lutz O, Jirgensons B (1931) New method for the grouping of optically active a-amino acids in the dextro-or levo-series. II. Abhandlungen 64:1221–1232
Acknowledgments
The authors gratefully acknowledge the funding of this research by the Ministry of Education and Science of Ukraine (Project #0116U001520), National Science Foundation (NSF/CREST HRD-1547754) and PREM (DMR-1205194) grants. This work also used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by the National Science Foundation (grant #ACI-1053575).
Author information
Authors and Affiliations
Corresponding authors
Additional information
This paper is dedicated to Peter Politzer as a recognition of his outstanding contributions to the field of quantum and computational chemistry on the occasion of his 80th birthday.
This paper belongs to Topical Collection P. Politzer 80th Birthday Festschrift.
Eugene Voronkov is an independent researcher.
Electronic supplementary material
ESM 1
(DOCX 112 kb)
Rights and permissions
About this article
Cite this article
Kapusta, K., Sizochenko, N., Karabulut, S. et al. QSPR modeling of optical rotation of amino acids using specific quantum chemical descriptors. J Mol Model 24, 59 (2018). https://doi.org/10.1007/s00894-018-3593-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-018-3593-z