Abstract
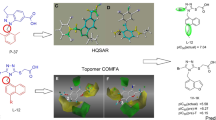
Human sodium-dependent glucose co-transporter 2 (hSGLT2) is a crucial therapeutic target in the treatment of type 2 diabetes. In this study, both comparative molecular field analysis (CoMFA) and comparative molecular similarity indices analysis (CoMSIA) were applied to generate three-dimensional quantitative structure–activity relationship (3D-QSAR) models. In the most accurate CoMFA-based and CoMSIA-based QSAR models, the cross-validated coefficients (r2cv) were 0.646 and 0.577, respectively, while the non-cross-validated coefficients (r2) were 0.997 and 0.991, respectively, indicating that both models were reliable. In addition, we constructed a homology model of hSGLT2 in the absence of a crystal structure. Molecular docking was performed to explore the bonding mode of inhibitors to the active site of hSGLT2. Molecular dynamics (MD) simulations and binding free energy calculations using MM-PBSA and MM-GBSA were carried out to further elucidate the interaction mechanism. With regards to binding affinity, we found that hydrogen-bond interactions of Asn51 and Glu75, located in the active site of hSGLT2, with compound 40 were critical. Hydrophobic and electrostatic interactions were shown to enhance activity, in agreement with the results obtained from docking and 3D-QSAR analysis. Our study results shed light on the interaction mode between inhibitors and hSGLT2 and may aid in the development of C-aryl glucoside SGLT2 inhibitors.







Similar content being viewed by others
References
American Diabetes Association (2014) Diagnosis and classification of diabetes mellitus. Diabetes Care 37(Suppl 1):S81–90
Bonner C, Kerr-Conte J, Gmyr V, Queniat G, Moerman E, Thevenet J, Beaucamps C, Delalleau N, Popescu I, Malaisse WJ, Sener A, Deprez B, Abderrahmani A, Staels B, Pattou F (2015) Inhibition of the glucose transporter SGLT2 with dapagliflozin in pancreatic alpha cells triggers glucagon secretion. Nat Med 21(5):512–U139. https://doi.org/10.1038/nm.3828
Tahrani AA, Bailey CJ, Del Prato S, Barnett AH (2011) Management of type 2 diabetes: new and future developments in treatment. Lancet 378(9786):182–197. https://doi.org/10.1016/S0140-6736(11)60207-9
Inzucchi SE (2002) Oral antihyperglycemic therapy for type 2 diabetes. Scientific review. JAMA J Am Med Assoc 287(3):360–372. https://doi.org/10.1001/jama.287.3.360
Tahrani AA, Piya MK, Kennedy A, Barnett AH (2010) Glycaemic control in type 2 diabetes: targets and new therapies. Pharmacol Ther 125(2):328–361. https://doi.org/10.1016/j.pharmthera.2009.11.001
Chao EC, Henry RR (2010) SGLT2 inhibition—a novel strategy for diabetes treatment. Nat Rev Drug Discov 9(7):551–559. https://doi.org/10.1038/nrd3180
Prentki M, Nolan CJ (2006) Islet β cell failure in type 2 diabetes. J Clin Invest 116(7):1802–1812. https://doi.org/10.1172/JCI29103
Armstrong K, Moye E, Williams S, Berlin JA, Reynolds EE (2007) Screening mammography in women 40 to 49 years of age: a systematic review for the American College of Physicians. Ann Intern Med 146(7):516–526. https://doi.org/10.7326/0003-4819-146-7-200704030-00008
Inzucchi SE, Bergenstal RM, Buse JB, Diamant M, Ferrannini E, Nauck M, Peters AL, Tsapas A, Wender R, Matthews DR (2012) Management of hyperglycaemia in type 2 diabetes, 2015: a patient-centred approach. Update to a position statement of the American Diabetes Association and the European Association for the Study of Diabetes. Diabetes Care 35(3):1364–1379
Yki-Jarvinen H (2004) Thiazolidinediones. N Engl J Med 351(11):1106–1118. https://doi.org/10.1056/NEJMra041001
Yao CH, Song JS, Chen CT, Yeh TK, Hung MS, Chang CC, Liu YW, Yuan MC, Hsieh CJ, Huang CY (2010) Discovery of novel N-beta-D-xylosylindole derivatives as sodium-dependent glucose cotransporter 2 (SGLT2) inhibitors for the management of hyperglycemia in diabetes. J Med Chem 54(1):166–178
Isaji M (2007) Sodium-glucose cotransporter inhibitors for diabetes. Curr Opin Invest Drugs (Thomson Sci) 8(4):285–292
Wright EM (2001) Renal Na(+)-glucose cotransporters. Am J Physiol Ren Physiol 280(1):F10–F18
Rahmoune H, Thompson PW, Ward JM, Smith CD, Hong G, Brown J (2005) Glucose transporters in human renal proximal tubular cells isolated from the urine of patients with non–insulin-dependent diabetes. Diabetes 54(12):3427–3434
Zhang W, Welihinda A, Mechanic J, Ding H, Zhu L, Lu Y, Deng Z, Sheng Z, Lv B, Chen Y, Roberge JY, Seed B, Wang YX (2011) EGT1442, a potent and selective SGLT2 inhibitor, attenuates blood glucose and HbA(1c) levels in db/db mice and prolongs the survival of stroke-prone rats. Pharmacol Res 63(4):284–293. https://doi.org/10.1016/j.phrs.2011.01.001
Idris I, Donnelly R (2009) Sodium–glucose co-transporter-2 inhibitors: an emerging new class of oral antidiabetic drug. Diabetes Obes Metab 11(2):79–88. https://doi.org/10.1111/j.1463-1326.2008.00982.x
Kumari B, Chetia D (2013) In-silico docking studies of selected n-glycoside bearing tetrazole ring in the treatment of hyperglycemia showing inhibitory activity on SGLT. Int J Pharm Pharm Sci 5:633–63818
Washburn WN (2012) Sodium glucose co-transporter 2 (SGLT2) inhibitors: novel antidiabetic agents. Expert Opin Ther Pat 22(5):483–494. https://doi.org/10.1517/13543776.2012.680437
Ehrenkranz JRL, Lewis NG, Kahn CR, Roth J (2005) Phlorizin: a review. Diabetes/Metab Res Rev 21(1):31–38. https://doi.org/10.1002/dmrr.532
Bailey CJ, Gross JL, Pieters A, Bastien A, List JF (2010) Effect of dapagliflozin in patients with type 2 diabetes who have inadequate glycaemic control with metformin: a randomised, double-blind, placebo-controlled trial. Lancet 375(9733):2223–2233. https://doi.org/10.1016/S0140-6736(10)60407-2
Ferrannini E, Ramos SJ, Salsali A, Tang W, List JF (2010) Dapagliflozin monotherapy in type 2 diabetic patients with inadequate glycemic control by diet and exercise: a randomized, double-blind, placebo-controlled, phase 3 trial. Diabetes Care 33(10):2217–2224
Rosenstock J, Vico M, Wei L, Salsali A, List JF (2012) Effects of dapagliflozin, an SGLT2 inhibitor, on HbA1c, body weight, and hypoglycemia risk in patients with type 2 diabetes inadequately controlled on pioglitazone monotherapy. Diabetes Care 35(7):1473–1478
Davis SN (2014) Canagliflozin versus glimepiride treatment in patients with type 2 diabetes inadequately controlled with metformin (CANTATA-SU trial). Expert Rev Clin Pharmacol 7(1):21–23. https://doi.org/10.1586/17512433.2014.864950
Häring H-U, Merker L, Seewaldt-Becker E, Weimer M, Meinicke T, Woerle HJ, Broedl UC, on behalf of the E-REGMTI (2013) Empagliflozin as add-on to metformin plus sulfonylurea in patients with type 2 diabetes: a 24-week, randomized, double-blind, placebo-controlled trial. Diabetes Care 36(11):3396–3404. https://doi.org/10.2337/dc12-2673
Bhongade BA, Gadad AK (2006) Insight into the structural requirements of urokinase-type plasminogen activator inhibitors based on 3D QSAR CoMFA/CoMSIA models. J Med Chem 49(2):475
Rodriguez R, Chinea G, Lopez N, Pons T, Vriend G (1998) Homology modeling, model and software evaluation: three related resources. Bioinformatics 14(6):523–528. https://doi.org/10.1093/bioinformatics/14.6.523
Udatha DB, Sugaya N, Olsson L, Panagiotou G (2012) How well do the substrates KISS the enzyme? Molecular docking program selection for feruloyl esterases. Sci Rep 2:323. https://doi.org/10.1038/srep00323
Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS, Olson AJ (2016) Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc 11(5):905–919
Vettoretti G, Moroni E, Sattin S, Tao J, Agard DA, Bernardi A, Colombo G (2016) Molecular dynamics simulations reveal the mechanisms of allosteric activation of Hsp90 by designed ligands. Sci Rep 6:23830. https://doi.org/10.1038/srep23830
Böhm M, Stürzebecher J, Klebe G (1999) Three-dimensional quantitative structure–activity relationship analyses using comparative molecular field analysis and comparative molecular similarity indices analysis to elucidate selectivity differences of inhibitors binding to trypsin, thrombin, and factor Xa. J Med Chem 42(3):458–477. https://doi.org/10.1021/jm981062r
Li ML, Ren YJ, Dong MH, Ren WX (2015) Design, synthesis and structural exploration of novel fluorinated dabigatran derivatives as direct thrombin inhibitors. Eur J Med Chem 96:122–138. https://doi.org/10.1016/j.ejmech.2015.04.012
Watanabe A, Choe S, Chaptal V, Rosenberg JM, Wright EM, Grabe M, Abramson J (2010) The mechanism of sodium and substrate release from the binding pocket of vSGLT. Nature 468(7326):988–991. https://doi.org/10.1038/nature09580
Tripos, Inc. (2006) Sybyl 7.3. Tripos, Inc., St. Louis
Golbraikh A, Tropsha A (2002) Beware of q2! J Mol Graph Model 20(4):269–276
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22(2):195–201. https://doi.org/10.1093/bioinformatics/bti770
Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31(13):3381–3385
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18(15):2714–2723. https://doi.org/10.1002/elps.1150181505
Sun YF, De Biasio F, Qiao HL, Iovinella I, Yang SX, Ling Y, Riviello L, Battaglia D, Falabella P, Yang XL, Pelosi P (2012) Two odorant-binding proteins mediate the behavioural response of aphids to the alarm pheromone (E)-β-farnesene and structural analogues. PLoS One 7(3):e32759. https://doi.org/10.1371/journal.pone.0032759
Grygorenko OO, Babenko P, Volochnyuk DM, Raievskyi O, Komarov IV (2016) Following Ramachandran: exit vector plots (EVP) as a tool to navigate chemical space covered by 3D bifunctional scaffolds. The case of cycloalkanes. RSC Adv 6(21):17595–17605
Bodade RG, Beedkar SD, Manwar AV, Khobragade CN (2010) Homology modeling and docking study of xanthine oxidase of Arthrobacter sp. XL26. Int J Biol Macromol 47(2):298–303. https://doi.org/10.1016/j.ijbiomac.2010.04.002
Jain AN (1996) Scoring noncovalent protein–ligand interactions: a continuous differentiable function tuned to compute binding affinities. J Comput Aided Mol Des 10(5):427–440
Welch W, Ruppert J, Jain AN (1996) Hammerhead: fast, fully automated docking of flexible ligands to protein binding sites. Chem Biol 3(6):449–462
Clark RD, Strizhev A, Leonard JM, Blake JF, Matthew JB (2002) Consensus scoring for ligand/protein interactions. J Mol Graph Model 20(4):281–295
Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P (2003) A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem 24(16):1999–2012. https://doi.org/10.1002/jcc.10349
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general Amber force field. J Comput Chem 25(9):1157–1174. https://doi.org/10.1002/jcc.20035
Darden T, York D, Pedersen L (1993) Particle mesh Ewald—an N.log(N) method for Ewald sums in large systems. J Chem Phys 98(12):10089–10092. https://doi.org/10.1063/1.464397
Ryckaert J-P, Ciccotti G, Berendsen HJC (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23(3):327–341. https://doi.org/10.1016/0021-9991(77)90098-5
Onufriev A, Bashford D, Case DA (2004) Exploring protein native states and large-scale conformational changes with a modified generalized Born model. Proteins 55(2):383–394. https://doi.org/10.1002/prot.20033
Weiser J, Shenkin PS, Still WC (2015) Approximate atomic surfaces from linear combinations of pairwise overlaps (LCPO). J Comput Chem 20(2):217–230
Lee J, Lee SH, Seo HJ, Son EJ, Lee SH, Jung ME, Lee M, Han HK, Kim J, Kang J, Lee J (2010) Novel C-aryl glucoside SGLT2 inhibitors as potential antidiabetic agents: 1,3,4-thiadiazolylmethylphenyl glucoside congeners. Bioorg Med Chem 18(6):2178–2194. https://doi.org/10.1016/j.bmc.2010.01.073
Jr JHW (1963) Hierarchical grouping to optimize an objective function. J Am Stat Assoc 58(301):236–244
Xu J, Yuan H, Ran T, Zhang Y, Liu H, Lu S, Xiong X, Xu A, Jiang Y, Lu T, Chen Y (2015) A selectivity study of sodium-dependent glucose cotransporter 2/sodium-dependent glucose cotransporter 1 inhibitors by molecular modeling. J Mol Recognit 28(8):467–479. https://doi.org/10.1002/jmr.2464
Roe DR, Cheatham 3rd TE (2013) PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput 9(7):3084–3095. https://doi.org/10.1021/ct400341p
Miller 3rd BR, McGee Jr TD, Swails JM, Homeyer N, Gohlke H, Roitberg AE (2012) MMPBSA.py: an efficient program for end-state free energy calculations. J Chem Theory Comput 8(9):3314–3321. https://doi.org/10.1021/ct300418h
Wang J, Zhou X, Liu S, Li G, Zhang B, Deng X, Niu X (2015) Novel inhibitor discovery and the conformational analysis of inhibitors of listeriolysin O via protein–ligand modeling. Sci Rep 5:8864. https://doi.org/10.1038/srep08864
Decherchi S, Berteotti A, Bottegoni G, Rocchia W, Cavalli A (2015) The ligand binding mechanism to purine nucleoside phosphorylase elucidated via molecular dynamics and machine learning. Nat Commun 6:6155. https://doi.org/10.1038/ncomms7155
Acknowledgements
This project was supported by the National Science Foundation for Fostering Talents in Basic Research of China (no. J1210064), the National Nature Science Foundation of China (21172257), and the National Science and Technology Pillar Program (2015BAK45B01) of China. All molecular dynamics simulation work was supported by Peking University. We would also like to thank Dr. Gu Jiangyong for helpful and productive discussions.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
ESM 1
(DOCX 4867 kb)
Rights and permissions
About this article
Cite this article
Dong, L., Feng, R., Bi, J. et al. Insight into the interaction mechanism of human SGLT2 with its inhibitors: 3D-QSAR studies, homology modeling, and molecular docking and molecular dynamics simulations. J Mol Model 24, 86 (2018). https://doi.org/10.1007/s00894-018-3582-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-018-3582-2