Abstract
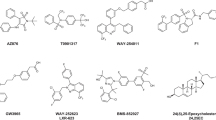
Liver X receptor (LXR), a member of the nuclear receptor superfamily, mainly serves as a reverse cholesterol transporter in lipid metabolism. It has been demonstrated that LXR is a promising target for the treatment of cardiovascular diseases. LXR is also involved in cancer metabolism, glucose homeostasis, immunity, and various physiological processes. The antitumor function of LXR has become of great interest to researchers in recent years. However, while it is believed that activating LXR with small molecules could be a promising approach to cancer treatment, effective drugs that target LXR are yet to be reported. To find compounds that are potentially capable of activating LXR, we utilized a high-throughput screening method to search the MolMall database for suitable compounds. Seven candidates with lower GB/SA Hawkins scores than the reference ligand T0901317 were identified. Based on the results of molecular dynamics (MD) simulations, binding free energy analysis, and an analysis of the agonism mechanism, ZINC90512020 and ZINC3845032 were predicted to have high affinities for LXR and high relative stabilization, and were therefore selected as potential LXR agonists. Both of these compounds will undergo further development with a view to utilizing them for the treatment of LXR-related cardiovascular diseases or cancers.




Similar content being viewed by others
References
Brown MS, Goldstein JL (1997) The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89(3):331
Stocker R, Keaney Jr JF (2004) Role of oxidative modifications in atherosclerosis. Physiol Rev 84(4):1381–1478
Steffensen KR (2015) Are synthetic compounds that silence the liver-X-receptor the next generation of anti-cancer drugs? Cancer Cell 28(1):3–4
Joseph SB, Tontonoz P (2003) LXRs: new therapeutic targets in atherosclerosis? Curr Opin Pharmacol 3(2):192–197
Phelan C et al (2008) Selective partial agonism of liver X receptor alpha is related to differential corepressor recruitment. Mol Endocrinol 22(10):2241–2249
Zhao CY, Dahlmanwright K (2010) Liver X receptor in cholesterol metabolism. J Endocrinol 204(3):233–240
Repa JJ et al (2000) Regulation of absorption and ABC1-mediated efflux of cholesterol by RXR heterodimers. Science 289(5484):1524
Klucken J et al (2000) ABCG1 (ABC8), the human homolog of the Drosophila white gene, is a regulator of macrophage cholesterol and phospholipid transport. Proc Natl Acad Sci USA 97(2):817
Laffitte BA et al (2001) LXRs control lipid-inducible expression of the apolipoprotein E gene in macrophages and adipocytes. Proc Natl Acad Sci 98(2):507–512
Berge KE et al (2000) Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent ABC transporters. Science 290(5497):1771–1775
Chiang JY, Kimmel R, Stroup D (2001) Regulation of cholesterol 7alpha-hydroxylase gene (CYP7A1) transcription by the liver orphan receptor (LXRalpha). Gene 262(1–2):257
Joseph SB et al (2002) Direct and indirect mechanisms for regulation of fatty acid synthase gene expression by liver X receptors. J Biol Chem 277(13):11019–11025
Mehrotra A, Kaul D, Joshi K (2011) LXR-α selectively reprogrammes cancer cells to enter into apoptosis. Mol Cell Biochem 349(1):41–55
Gabbi C, Gustafsson JÅ (2010) Estrogen-dependent gallbladder carcinogenesis in LXRβ−/− female mice. Proc Natl Acad Sci USA 107(33):14763–14768
Vedin LL et al (2009) The oxysterol receptor LXR inhibits proliferation of human breast cancer cells. Carcinogenesis 30(4):575–579
Bovenga F, Moschetta A (2015) Uncoupling nuclear receptor LXR and cholesterol metabolism in cancer. Cell Metab 21(4):517
Flaveny CA et al (2015) Broad anti-tumor activity of a small molecule that selectively targets the Warburg effect and lipogenesis. Cancer Cells 28(1):42
Irwin JJ, Shoichet BK (2004) ZINC—a free database of commercially available compounds for virtual screening. J Chem Inf Model 45(1):177–182
Xu HL et al (2014) In silico identification of novel kinase inhibitors targeting wild-type and T315I mutant ABL1 from FDA-approved drugs. Mol BioSyst 10(6):1524
Ceretomassagué A et al (2012) DecoyFinder: an easy-to-use Python GUI application for building target-specific decoy sets. Bioinformatics 28(12):1661
Robin X et al (2011) pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinf 12(1):77
Pettersen EF et al (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25(13):1605
Lang PT et al (2009) DOCK 6: combining techniques to model RNA–small molecule complexes. RNA 15(6):1219–1230
Yu QJ et al (2011) In silico analysis of molecular mechanisms of Galanthus nivalis agglutinin-related lectin-induced cancer cell death from carbohydrate-binding motif evolution hypothesis. Appl Biochem Biotechnol 165(3):1037–1046
Liu H, Kuntz ID, Zou X (2004) Pairwise GB/SA scoring function for structure-based drug design. J Phys Chem B 108(17):5453–5462
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7:42717
Pronk S et al (2013) GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29(7):845–854
Hornak V et al (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins Struct Funct Bioinf 65(3):712–725
Jorgensen WL et al (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79(2):926–935
Berendsen HJC, Marrink SJ (1993) Molecular dynamics of water transport through membranes: water from solvent to solute. Pure Appl Chem 65(12):2513–2520
Hess B et al (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18(12):1463–1472
Miyamoto S, Kollman PA (1992) Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J Comput Chem 13(8):952–962
Essmann U et al (1995) A smooth particle mesh Ewald method. J Chem Phys 103(19):8577
Homeyer N, Gohlke H (2012) Free energy calculations by the molecular mechanics Poisson−Boltzmann surface area method. Molecular Inf 31(2):114–122
Kollman PA et al (2000) Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res 33(12):889–897
Honig B, Nicholls A (1995) Classical electrostatics in biology and chemistry. Science 268(5214):1144–1149
Still WC et al (1990) Semianalytical treatment of solvation for molecular mechanics and dynamics. J Am Chem Soc 112(16):6127–6129
Wan Z et al (2015) Computer-assisted identification of novel small molecule inhibitors targeting GLUT1. J Mol Struct 1101:57–65
Kumari R, Kumar R, Lynn A (2014) g_mmpbsa—a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model 54(7):1951–1962
Wallace AC, Thornton JM, Laskowski RA (1995) LIGPLOT: a program to generate schematic diagrams of protein–ligand interactions. Protein Eng 8(2):127–134
Schneidman-Duhovny D et al (2008) PharmaGist: a webserver for ligand-based pharmacophore detection. Nucleic Acids Res 36:W223–W228
Zhang W et al (2016) Computer-aided identification of potential TYK2 inhibitors from drug database. J Mol Struct 1122:309–317
Lipinski CA (2004) Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol 1(4):337–341
Sim WC et al (2015) Cinnamamides, novel liver X receptor antagonists that inhibit ligand-induced lipogenesis and fatty liver. J Pharmacol Exp Ther 355(3):362
Stachel SJ et al (2016) Identification and in vivo evaluation of liver X receptor β (LXRβ) selective agonists for the potential treatment of Alzheimer's disease. J Med Chem 59(7):3489–3498
Janowski BA et al (1996) An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature 383(6602):728–731
Lehmann JM et al (1997) Activation of the nuclear receptor LXR by oxysterols defines a new hormone response pathway. J Biol Chem 272(6):3137–3140
Collins JL et al (2002) Identification of a nonsteroidal liver X receptor agonist through parallel array synthesis of tertiary amines. J Med Chem 45(10):1963–1966
Schultz JR et al (2000) Role of LXRs in control of lipogenesis. Genes Dev 14(22):2831–2838
Candelaria NR et al (2014) Antiproliferative effects and mechanisms of liver X receptor ligands in pancreatic ductal adenocarcinoma cells. PLoS One 9(9):e106289
Manglik A et al (2016) Structure-based discovery of opioid analgesics with reduced side effects. Nature 537(7619):1
Li P et al (2013) The gating charge pathway of an epilepsy-associated potassium channel accommodates chemical ligands. Cell Res 23(9):1106
Schneidman-Duhovny D et al (2008) PharmaGist: a webserver for ligand-based pharmacophore detection. Nucleic Acids Res 36:W223
Hoerer S et al (2003) Crystal structure of the human liver X receptor β ligand-binding domain in complex with a synthetic agonist. J Mol Biol 334(5):853–861
Acknowledgments
We are grateful to our colleagues for their critical reviews and constructive suggestions regarding this manuscript. This work was supported in part by grants from the National Natural Science Foundation of China (nos. 81373311, 31300674, 81173093, 30970643, and J1103518)
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Fig. S1
Backbone RMSDs from the second MD simulations of the LXR–ligand complexes. (GIF 248 kb)
Fig. S2
Backbone RMSDs from the third MD simulations of the LXR–ligand complexes. (GIF 224 kb)
Fig. S3
RMSF profiles for the protein backbones of the LXR–ligand complexes based on the last 10 ns (20–30 ns) of the second MD simulation of each complex. (GIF 52 kb)
Fig. S4
RMSF profiles for the protein backbones of the LXR–ligand complexes based on the last 10 ns (20–30 ns) of the third MD simulation of each complex (GIF 51 kb)
Table S1
(DOCX 26 kb)
Table S2
(DOC 92 kb)
Table S3
(DOC 37 kb)
Rights and permissions
About this article
Cite this article
Wang, X., Lu, K., Luo, H. et al. In silico identification of small molecules as novel LXR agonists for the treatment of cardiovascular disease and cancer. J Mol Model 24, 57 (2018). https://doi.org/10.1007/s00894-018-3578-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-018-3578-y