Abstract
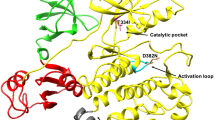
The deregulated breakpoint cluster region (Bcr)–Abelson tyrosine kinase (Abl) fusion protein represents an attractive pharmacological target for the treatment of chronic myeloid leukemia (CML). The high affinity of monobody AS25 was designed to target the Src homology 2 (SH2) domain of Bcr-Abl, leading to allosteric inhibition of Bcr-Abl through formation of protein–protein interactions. An I164E mutation in the SH2 domain disrupts AS25 binding to the SH2 domain of Bcr-Abl. The detailed mechanisms, however, remain to be unresolved. Here, molecular dynamics (MD) simulations and binding free energy calculations were performed to explore the conformational and energetic differences between the wild-type (WT) complexes of Bcr-Abl SH2 domain and AS25 (SH2WT–AS25) as well as the mutated complexes (SH2I164E–AS25). The results revealed that I164E mutation not only caused an increase in the conformational flexibility of SH2–AS25 complexes, but also weakened the binding affinity of AS25 to SH2. The comparative binding modes of SH2-AS25 complexes between WT and the I164E mutant were comprehensively analyzed to unravel the disruption of hydrophobic and hydrogen bonding interactions in the interface of the SH2-AS25 complex triggered by the I164E mutation. The results obtained may help to design the next generation of higher affinity Bcr-Abl SH2-specific peptide inhibitors.







Similar content being viewed by others
References
Cilloni D, Saglio G (2012) Molecular pathways: BCR-ABL. Clin Cancer Res 18:930–937
Lambert GK, Duhme-Klair A-K, Morgan T, Ramjee MK (2013) The background, discovery and clinical development of BCR-ABL inhibitors. Drug Discov Today 18:992–1000
Balabanov S, Braig M, Brümmendorf TH (2014) Current aspects in resistance against tyrosine kinase inhibitors in chronic myelogenous leukemia. Drug Discov Today Technol 11:89–99
Desogus S, Schenone S, Brullo C et al (2015) Bcr-Abl tyrosine kinase inhibitors: a patent review. Expert Opin Ther Pat 25:397–412
Roskoski Jr R (2015) A historical overview of protein kinases and their targeted small molecule inhibitors. Pharmacol Res 100:1–23
Skora L, Mestan J, Fabbro D et al (2013) NMR reveals the allosteric opening and closing of Abelson tyrosine kinase by ATP-site and myristoyl pocket inhibitors. Proc Natl Acad Sci USA 110:E4437–E4445
Pemovska T, Johnson E, Kontro M et al (2015) Axitinib effectively inhibits BCR-ABL1(T315I) with a distinct binding conformation. Nature 519:102–105
Yang K, Fu L (2015) Mechanisms of resistance to BCR-ABL TKIs and the therapeutic strategies: a review. Crit Rev Oncol Hematol 93:277–292
Zhang J, Adrián FJ, Jahnke W et al (2010) Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors. Nature 463:501–506
Lu S, Jang H, Muratcioglu S et al (2016) Ras conformational ensembles, Allostery, and signaling. Chem Rev 116:6607–6665
Lu S, Jang H, Gu S, et al (2016) Drugging Ras GTPase: a comprehensive mechanistic and signaling structural view. Chem Soc Rev 45:4929–4952
Tanaka T, Williams RL, Rabbitts TH (2007) Tumour prevention by a single antibody domain targeting the interaction of signal transduction proteins with RAS. EMBO J 26:3250–3259
Salzman GS, Ackerman SD, Ding C et al (2016) Structural basis for regulation of GPR56/ADGRG1 by its alternatively spliced extracellular domains. Neuron 91:1292–1304
Wojcik J, Lamontanara AJ, Grabe G et al (2016) Allosteric inhibition of Bcr-Abl kinase by high affinity Monobody inhibitors directed to the Src homology 2 (SH2)-kinase Interface. J Biol Chem 291:8836–8847
Case DA, Cheatham TE, Darden T et al (2005) The Amber biomolecular simulation programs. J Comput Chem 26:1668–1688
Hornak V, Abel R, Okur A et al (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65:712–725
Jorgensen WL, Chandrasekhar J, Madura JD et al (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926
Lu S, Huang W, Wang Q et al (2014) The structural basis of ATP as an allosteric modulator. PLoS Comput Biol 10:e1003831
Lu S, Deng R, Jiang H et al (2015) The mechanism of ATP-dependent allosteric protection of Akt kinase phosphorylation. Structure 23:1725–1734
Lu S, Banerjee A, Jang H et al (2015) GTP binding and oncogenic mutations may attenuate hypervariable region (HVR)-catalytic domain interactions in small GTPase K-Ras4B, exposing the effector binding site. J Biol Chem 290:28887–28900
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an N log(N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092
Ryckaert J-P, Ciccotti G, Berendsen HJC (1977) Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23:327–341
Hou T, Wang J, Li Y, Wang W (2011) Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model 51:69–82
Hou T, Wang J, Li Y, Wang W (2011) Assessing the performance of the molecular mechanics/Poisson Boltzmann surface area and molecular mechanics/generalized born surface area methods. II. The accuracy of ranking poses generated from docking. J Comput Chem 32:866–877
Ning L, Wang Q, Zheng Y et al (2015) Effects of the A117V mutation on the folding and aggregation of palindromic sequences (PrP113-120) in prion: insights from replica exchange molecular dynamics simulations. Mol BioSyst 11:647–655
Wang J, Morin P, Wang W et al (2001) Use of MM-PBSA in reproducing the binding free energies to HIV-1 RT of TIBO derivatives and predicting the binding mode to HIV-1 RT of efavirenz by docking and MM-PBSA. J Am Chem Soc 123:5221–5230
Kollman PA, Massova I, Reyes C et al (2000) Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res 33:889–897
Kong X, Pan P, Li D et al (2015) Importance of protein flexibility in ranking inhibitor affinities: modeling the binding mechanisms of piperidine carboxamides as type I1/2 ALK inhibitors. Phys Chem Chem Phys 17:6098–6113
Zhou Y, Zhang N, Chen W et al (2016) Underlying mechanisms of cyclic peptide inhibitors interrupting the interaction of CK2α/CK2β: comparative molecular dynamics simulation studies. Phys Chem Chem Phys 18:9202–9210
Yang B, Zhang H, Wang H (2015) Atomistic insights into the lung cancer-associated L755P mutation in HER2 resistance to lapatinib: a molecular dynamics study. J Mol Model 21:2580
Ni Z, Zhang T-C (2015) Computationally unraveling how ceritinib overcomes drug-resistance mutations in ALK-rearranged lung cancer. J Mol Model 21:175
Lang EJM, Heyes LC, Jameson GB, Parker EJ (2016) Calculated pKa variations expose dynamic allosteric communication networks. J Am Chem Soc 138:2036–2045
Guo X-Y, Qi R-P, Xu D-G et al (2015) Structural and energetic insight into the interactions between the benzolactam inhibitors and tumor marker HSP90α. Comput Biol Chem 58:182–191
Panjarian S, Iacob RE, Chen S et al (2013) Structure and dynamic regulation of Abl kinases. J Biol Chem 288:5443–5450
Nagar B, Hantschel O, Young MA et al (2003) Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell 112:859–871
Nagar B, Hantschel O, Seeliger M et al (2006) Organization of the SH3-SH2 unit in active and inactive forms of the c-Abl tyrosine kinase. Mol Cell 21:787–798
Jang H, Banerjee A, Chavan TS et al (2016) The higher level of complexity of K-Ras4B activation at the membrane. FASEB J 30:1643–1655
Lu S, Jang H, Nussinov R et al (2016) The structural basis of oncogenic mutations G12, G13 and Q61 in small GTPase K-Ras4B. Sci Rep 6:21949
Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797
Acknowledgment
The authors thank the high performance supercomputer center at Shanghai.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Funding
This work was supported by Shanghai Health and Family Planning Commission (20154Y0058).
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Mingfei Ji, Guodong Zheng, and Xiaolong Li wish it to be known that, in their opinion, the first three authors should be regarded as joint First Authors
Rights and permissions
About this article
Cite this article
Ji, M., Zheng, G., Li, X. et al. Computational dissection of allosteric inhibition of the SH2 domain of Bcr-Abl kinase by the monobody inhibitor AS25. J Mol Model 23, 183 (2017). https://doi.org/10.1007/s00894-017-3353-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-017-3353-5