Abstract
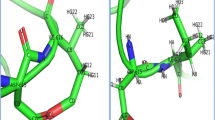
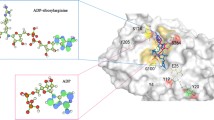
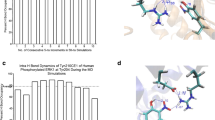
Homology modeling of the reductase domain of endothelial nitric oxide synthase (eNOS), which regulates the catalytic activity of eNOS, and molecular dynamics studies focusing especially on the serine residues S615, S633, and S1177 were performed. MD analysis of this structure revealed that S633 is highly flexible and accessible to solvent molecules, while S1177 becomes highly flexible when S633 is phosphorylated. The presence of intramolecular interactions between S1177 among the major serine residues underscores its structural importance to the efficient synthesis of nitric oxide in endothelium. In order to evaluate the appropriateness of phosphomimetic (for phosphorylation) and phosphomutant (for dephosphorylation) eNOSs for use as experimental model systems, the structural dynamics and conformational changes in phosphomimetic (S615D, S633D, S1177D) and phosphomutant (S615A, S633A, S1177A) eNOSs were investigated. Phosphomimetic and phosphomutant eNOSs portrayed S633 as a modulator of S1177, whereas such correlations could not be observed in native and phosphorylated eNOSs. Computational analysis of the docked complex revealed that phosphorylated pS1177 and pS615 have high affinity for Akt (one of the key kinases in the eNOS activation pathway), with a significant number of hydrogen bonds and salt bridges observed between these residues and Akt . This work therefore provides evidence of the subtle structural changes that occur within the reductase domain which contribute to the stability–flexibility–activity relationship of eNOS. Such subtle changes are of great importance in the context of regulated nitric oxide release by different phosphorylated forms of eNOS and the need to account for the existence of subtle differences between real proteins and experimental model systems.






Similar content being viewed by others
References
Moncada S, Higgs EA (2006) Nitric oxide and the vascular endothelium. Handb Exp Pharmacol 176:213–254
Förstermann U, Pollock JS, Nakane M (1993) Nitric oxide synthases in the cardiovascular system. Trends Cardiovasc Med 3:104–110
Bredt DS, Hwang PM, Snyder SH (1990) Localization of nitric oxide synthase indicating a neural role for nitric oxide. Nature 347:768–770
Wright CD, Mülsch A, Busse R, Osswald H (1989) Generation of nitric oxide by human neutrophils. Biochem Biophys Res Commun 160:813–819
Janssens SP, Shimouchi A, Quertermous T, Bloch DB, Bloch KD (1992) Cloning and expression of a cDNA encoding human endothelium-derived relaxing factor/nitric oxide synthase. J Biol Chem 267:14519–14522
Alderton WK, Cooper CE, Knowles RG (2001) Nitric oxide synthases: structure, function and inhibition. Biochem J 357:593–615
Cines DB, Pollak ES, Buck CA, Loscalzo J, Zimmerman GA et al (1998) Endothelial cells in physiology and in the pathophysiology of vascular disorders. Blood 91:3527–3561
Cooke JP, Dzau VJ (1997) Nitric oxide synthase: role in the genesis of vascular disease. Annu Rev Med 48:489–509
Du M, Yeh HC, Berka V, Wang LH, Tsai A (2003) Redox properties of human endothelial nitric-oxide synthase oxygenase and reductase domains purified from yeast expression system. J Biol Chem 278:6002–6011
Martásek P, Miller RT, Liu Q, Roman LJ, Salerno JC et al (1998) The C331A mutant of neuronal nitric-oxide synthase is defective in arginine binding. J Biol Chem 273:34799–34805
Sessa WC, Harrison JK, Barber CM, Zeng D, Durieux ME et al (1992) Molecular cloning and expression of a cDNA encoding endothelial cell nitric oxide synthase. J Biol Chem 267:15274–15276
Feron O, Belhassen L, Kobzik L, Smith TW, Kelly RA et al (1996) Endothelial nitric oxide synthase targeting to caveolae. Specific interactions with caveolin isoforms in cardiac myocytes and endothelial cells. J Biol Chem 271:22810–22814
Mount PF, Kemp BE, Power DA (2007) Regulation of endothelial and myocardial NO synthesis by multi-site eNOS phosphorylation. J Mol Cell Cardiol 42:271–279
McCabe TJ, Fulton D, Roman LJ, Sessa WC (2000) Enhanced electron flux and reduced calmodulin dissociation may explain “calcium-independent” eNOS activation by phosphorylation. J Biol Chem 275:6123–6128
Morales-Ruiz M, Fulton D, Sowa G, Languino LR, Fujio Y et al (2000) Vascular endothelial growth factor-stimulated actin reorganization and migration of endothelial cells is regulated via the serine/threonine kinase Akt. Circ Res 86:892–896
Michell BJ, Z-p C, Tiganis T, Stapleton D, Katsis F et al (2001) Coordinated control of endothelial nitric-oxide synthase phosphorylation by protein kinase C and the cAMP-dependent protein kinase. J Biol Chem 276:17625–17628
Lane P, Gross SS (2002) Disabling a C-terminal autoinhibitory control element in endothelial nitric-oxide synthase by phosphorylation provides a molecular explanation for activation of vascular NO synthesis by diverse physiological stimuli. J Biol Chem 277:19087–19094
Boo YC, Sorescu G, Boyd N, Shiojima I, Walsh K et al (2002) Shear stress stimulates phosphorylation of endothelial nitric-oxide synthase at Ser1179 by Akt-independent mechanisms: role of protein kinase A. J Biol Chem 277:3388–3396
Harris MB, Ju H, Venema VJ, Liang H, Zou R et al (2001) Reciprocal phosphorylation and regulation of endothelial nitric-oxide synthase in response to bradykinin stimulation. J Biol Chem 276:16587–16591
Montagnani M, Chen H, Barr VA, Quon MJ (2001) Insulin-stimulated activation of eNOS is independent of Ca2+ but requires phosphorylation by Akt at Ser (1179). J Biol Chem 276:30392–30398
Michell BJ, Griffiths JE, Mitchelhill KI, Rodriguez-Crespo I, Tiganis T et al (1999) The Akt kinase signals directly to endothelial nitric oxide synthase. Curr Biol 9:845–848
Rafikov R, Fonseca FV, Kumar S, Pardo D, Darragh C et al (2011) eNOS activation and NO function: structural motifs responsible for the posttranslational control of endothelial nitric oxide synthase activity. J Endocrinol 210:271–284
Boo YC, Sorescu GP, Bauer PM, Fulton D, Kemp BE et al (2003) Endothelial NO synthase phosphorylated at SER635 produces NO without requiring intracellular calcium increase. Free Radic Biol Med 35:729–741
Fischmann TO, Hruza A, Niu XD, Fossetta JD, Lunn CA et al (1999) Structural characterization of nitric oxide synthase isoforms reveals striking active-site conservation. Nat Struct Biol 6:233–242
Garcin ED, Bruns CM, Lloyd SJ, Hosfield DJ, Tiso M et al (2004) Structural basis for isozyme-specific regulation of electron transfer in nitric-oxide synthase. J Biol Chem 279:37918–37927
Atochin DN, Wang A, Liu VW, Critchlow JD, Dantas AP et al (2007) The phosphorylation state of eNOS modulates vascular reactivity and outcome of cerebral ischemia in vivo. J Clin Invest 117:1961–1967
Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D et al. (2006) Comparative protein structure modeling using Modeller. Curr Protocol Bioinform Unit 5.6. Wiley, Hoboken
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 26:283–291
Lüthy R, Bowie JU, Eisenberg D (1992) Assessment of protein models with three-dimensional profiles. Nature 356:83–85
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2:1511–1519
Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE et al (2005) GROMACS: fast, flexible, and free. J Comput Chem 26:1701–1718
Margreitter C, Petrov D, Zagrovic B (2013) Vienna-PTM web server: a toolkit for MD simulations of protein post-translational modifications. Nucleic Acids Res W422–6
Martonák R, Laio A, Parrinello M (2003) Predicting crystal structures: the Parrinello–Rahman method revisited. Phys Rev Lett 90:075503
Cuendet MA, van Gunsteren WF (2007) On the calculation of velocity-dependent properties in molecular dynamics simulations using the leapfrog integration algorithm. J Chem Phys 127:184102
Ahmad S, Gromiha M, Fawareh H, Sarai A (2004) ASAView: database and tool for solvent accessibility representation in proteins. BMC Bioinform 1:5–51
de Vries SJ, van Dijk M, Bonvin AM (2010) The HADDOCK web server for data-driven biomolecular docking. Nat Protoc 5:883–897
de Vries SJ, Bonvin AM (2011) CPORT: a consensus interface predictor and its performance in prediction-driven docking with HADDOCK. PLoS One 6:e17695
Costantini S, Colonna G, Facchiano AM (2008) ESBRI: a web server for evaluating salt bridges in proteins. Bioinformation 3:137–138
Tina KG, Bhadra R, Srinivasan N (2007) PIC: Protein Interactions Calculator. Nucleic Acids Res 35:W473–W476
Bauer PM, Fulton D, Boo YC, Sorescu GP, Kemp BE (2003) Compensatory phosphorylation and protein-protein interactions revealed by loss of function and gain of function mutants of multiple serine phosphorylation sites in endothelial nitric-oxide synthase. J Biol Chem 278:14841–14849
Chen Z-P, Mitchelhill KI, Michell BJ, Stapleton D, Rodriguez-Crespo I et al (1999) AMP-activated protein kinase phosphorylation of endothelial NO synthase. FEBS Lett 443:285–289
Fulton D, Gratton JP, Sessa WC (2001) Post-translational control of endothelial nitric oxide synthase: why isn’t calcium/calmodulin enough? J Pharmacol Exp Ther 299:818–824
Chen Z, Peng IC, Sun W, Su MI, Hsu PH et al (2009) AMP-activated protein kinase functionally phosphorylates endothelial nitric oxide synthase Ser633. Circ Res 104:496–505
Dimmeler S, Fleming I, Fisslthaler B, Hermann C, Busse R et al (1999) Activation of nitric oxide synthase in endothelial cells by Akt-dependent phosphorylation. Nature 399:601–605
Fulton D, Gratton JP, McCabe TJ, Fontana J, Fujio Y et al (1999) Regulation of endothelium-derived nitric oxide production by the protein kinase Akt. Nature 399:597–601
Brouet A, Sonveaux P, Dessy C, Moniotte S, Balligand JL et al (2001) Hsp90 and caveolin are key targets for the proangiogenic nitric oxide-mediated effects of statins. Circ Res 89:866–873
Acknowledgments
We acknowledge Dr. M. Suresh Kumar for critical suggestions and encouragement and Mr. Om Prakash Sharma for help with the software. The Centre for Green Energy Technology is acknowledged for allowing N.T.D. to use their facilities as a visiting scholar. This work was partially supported by the Department of Biotechnology, Government of India, grant no. BT/PR7628/BRB/2006.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 5984 kb)
Rights and permissions
About this article
Cite this article
Devika, N.T., Amresh, P., Hassan, M. et al. Molecular modeling and simulation of the human eNOS reductase domain, an enzyme involved in the release of vascular nitric oxide. J Mol Model 20, 2470 (2014). https://doi.org/10.1007/s00894-014-2470-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-014-2470-7