Abstract
The present benchmarking study utilizes the RNA123 program for de novo prediction of tertiary structures of a set of 50 RNA molecules for which X-ray/NMR structures are available, based on the nucleic acid sequence only. All molecules contain a hairpin loop motif and a helical structure of canonical and non-canonical base pairs, interrupted by bulges and internal loops to various degrees. RNA molecules with double helices made up purely by canonical base pairing, and molecules containing symmetric internal loops of non-canonical base pairing are, overall, very well predicted. Structures containing bulges and asymmetric internal loops, and more complex structures containing multiple bulges and internal loops in the same molecule, result in larger deviations from their X-ray/NMR predicted structures due to higher degree of flexibility of the nucleotide bases in these regions. In a majority of the molecules included herein, the RNA123 program was, however, able to predict the tertiary structure with a heavy atom RMSD of less than 5 Å to the X-ray/NMR structure, and the models were in most cases structurally closer to the X-ray/NMR structures than models predicted by MC-Fold and MC-Sym. A set of RNA molecules containing pseudoknot tertiary structure motifs were included, but neither of the programs was able to predict the folding of the single-stranded stem onto the helix without additional structural input. The RNA123 program was then applied to predict the tertiary structure of the RNA segment of Macugen®, the first RNA aptamer approved for clinical use, and for which no tertiary structure has yet been solved. Four possible tertiary structures were predicted for this 27-nucleic-acid-long RNA molecule, which will be used in constructing a full model of the PEGylated aptamer and its interaction with the vascular endothelial growth factor target.

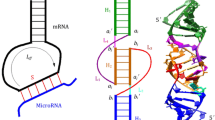
RNA123-predicted secondary and tertiary structures of RNA molecule containing a short helix with a hairpin loop. The predicted model is superposed with a structure determined by NMR.





Similar content being viewed by others
References
Gautheret D, Konings D, Gutell RR (1995) GU base pairing motifs in ribosomal RNA. RNA 1(8):807–814
Crick FHC (1966) Codon-anticodon pairing: the wobble hypothesis. J Mol Biol 19:548–555
Jaeger JA, Turner DH, Zuker M (1989) Improved predictions of secondary structures for RNA. Proc Natl Acad Sci USA 86(20):7706–7710
Mathews DH, Sabina J, Zuker M, Turner DH (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol 288(5):911–940. doi:10.1006/jmbi.1999.2700
Strazewski P, Biala E, Gabriel K, McClain WH (1999) The relationship of thermodynamic stability at a G x U recognition site to tRNA aminoacylation specificity. RNA 5(11):1490–1494
Doudna JA, Cormack BP, Szostak JW (1989) RNA structure, not sequence, determines the 5′ splice-site specificity of a group I intron. Proc Natl Acad Sci USA 86(19):7402–7406
Hur M, Waring RB (1995) Two group I introns with a C.G. basepair at the 5′ splice-site instead of the very highly conserved U.G. basepair: is selection post-translational? Nucleic Acids Res 23(21):4466–4470
Ban N, Nissen P, Hansen J, Moore PB, Steitz TA (2000) The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289(5481):905–920
Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, Yonath A (2000) Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell 102(5):615–623
Wimberly BT, Brodersen DE, Clemons WM Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V (2000) Structure of the 30S ribosomal subunit. Nature 407(6802):327–339. doi:10.1038/35030006
Abu Almakarem AS, Petrov AI, Stombaugh J, Zirbel CL, Leontis NB (2012) Comprehensive survey and geometric classification of base triples in RNA structures. Nucleic Acids Res 40(4):1407–1423. doi:10.1093/nar/gkr810
Agarwal T, Jayaraj G, Pandey SP, Agarwala P, Maiti S (2012) RNA G-quadruplexes: G-quadruplexes with “U” turns. Curr Pharm Des 18(14):2102–2111
Leontis NB, Westhof E (2001) Geometric nomenclature and classification of RNA base pairs. RNA 7(4):499–512
Woese CR, Winker S, Gutell RR (1990) Architecture of ribosomal RNA: constraints on the sequence of “tetra-loops”. Proc Natl Acad Sci USA 87(21):8467–8471
Heus HA, Pardi A (1991) Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science 253(5016):191–194
Baeyens KJ, De Bondt HL, Pardi A, Holbrook SR (1996) A curved RNA helix incorporating an internal loop with G.A. and A.A. non-Watson-Crick base pairing. Proc Natl Acad Sci USA 93(23):12851–12855
Carter RJ, Baeyens KJ, SantaLucia J, Turner DH, Holbrook SR (1997) The crystal structure of an RNA oligomer incorporating tandem adenosine-inosine mismatches. Nucleic Acids Res 25(20):4117–4122
Chen G, Znosko BM, Kennedy SD, Krugh TR, Turner DH (2005) Solution structure of an RNA internal loop with three consecutive sheared GA pairs. Biochemistry 44(8):2845–2856. doi:10.1021/bi048079y
Hammond NB, Tolbert BS, Kierzek R, Turner DH, Kennedy SD (2010) RNA internal loops with tandem AG pairs: the structure of the 5′GAGU/3′UGAG loop can be dramatically different from others, including 5′AAGU/3′UGAA. Biochemistry 49(27):5817–5827. doi:10.1021/bi100332r
Klein DJ, Schmeing TM, Moore PB, Steitz TA (2001) The kink-turn: a new RNA secondary structure motif. EMBO J 20(15):4214–4221. doi:10.1093/emboj/20.15.4214
Szep S, Wang J, Moore PB (2003) The crystal structure of a 26-nucleotide RNA containing a hook-turn. RNA 9(1):44–51
Rietveld K, Van Poelgeest R, Pleij CW, Van Boom JH, Bosch L (1982) The tRNA-like structure at the 3′ terminus of turnip yellow mosaic virus RNA. Differences and similarities with canonical tRNA. Nucleic Acids Res 10(6):1929–1946
Giedroc DP, Cornish PV (2009) Frameshifting RNA pseudoknots: structure and mechanism. Virus Res 139(2):193–208. doi:10.1016/j.virusres.2008.06.008
Gardner PP, Giegerich R (2004) A comprehensive comparison of comparative RNA structure prediction approaches. BMC Bioinforma 5:140. doi:10.1186/1471-2105-5-140
Parisien M, Major F (2008) The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature 452(7183):51–55. doi:10.1038/nature06684
Jossinet F, Ludwig TE, Westhof E (2010) Assemble: an interactive graphical tool to analyze and build RNA architectures at the 2D and 3D levels. Bioinformatics 26(16):2057–2059. doi:10.1093/bioinformatics/btq321
Martinez HM, Maizel JV Jr, Shapiro BA (2008) RNA2D3D: a program for generating, viewing, and comparing 3-dimensional models of RNA. J Biomol Struct Dyn 25(6):669–683. doi:10.1080/07391102.2008.10531240
Das R, Karanicolas J, Baker D (2010) Atomic accuracy in predicting and designing noncanonical RNA structure. Nat Methods 7(4):291–294. doi:10.1038/nmeth.1433
Tan RK, Petrov AS, Harvey SC (2006) YUP: a molecular simulation program for coarse-grained and multi-scaled models. J Chem Theory Comput 2(3):529–540. doi:10.1021/ct050323r
Sharma S, Ding F, Dokholyan NV (2008) iFoldRNA: three-dimensional RNA structure prediction and folding. Bioinformatics 24(17):1951–1952. doi:10.1093/bioinformatics/btn328
Ding F, Sharma S, Chalasani P, Demidov VV, Broude NE, Dokholyan NV (2008) Ab initio RNA folding by discrete molecular dynamics: from structure prediction to folding mechanisms. RNA 14(6):1164–1173. doi:10.1261/rna.894608
Jonikas MA, Radmer RJ, Laederach A, Das R, Pearlman S, Herschlag D, Altman RB (2009) Coarse-grained modeling of large RNA molecules with knowledge-based potentials and structural filters. RNA 15(2):189–199. doi:10.1261/rna.1270809
Sijenyi F, Saro P, Ouyang Z, Damm-Ganamet K, Wood M, Jiang J, SantaLucia J Jr (2011) The RNA folding problems: different levels of sRNA structure prediction. In: Leontis N, Westhof E (eds) RNA 3D structure analysis and prediction. Springer, Berlin, pp 91–117
Darty K, Denise A, Ponty Y (2009) VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 25(15):1974–1975. doi:10.1093/bioinformatics/btp250
Krieger E, Darden T, Nabuurs SB, Finkelstein A, Vriend G (2004) Making optimal use of empirical energy functions: force-field parameterization in crystal space. Proteins 57(4):678–683. doi:10.1002/prot.20251
Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P (2003) A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem 24(16):1999–2012. doi:10.1002/jcc.10349
Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) A smooth particle mesh Ewald method. J Chem Phys 103(19):8577–8593
Dallas A, Moore PB (1997) The loop E-loop D region of Escherichia coli 5S rRNA: the solution structure reveals an unusual loop that may be important for binding ribosomal proteins. Structure 5(12):1639–1653
Kitamura A, Muto Y, Watanabe S, Kim I, Ito T, Nishiya Y, Sakamoto K, Ohtsuki T, Kawai G, Watanabe K, Hosono K, Takaku H, Katoh E, Yamazaki T, Inoue T, Yokoyama S (2002) Solution structure of an RNA fragment with the P7/P9.0 region and the 3′-terminal guanosine of the tetrahymena group I intron. RNA 8(4):440–451
Zhang L, Doudna JA (2002) Structural insights into group II intron catalysis and branch-site selection. Science 295(5562):2084–2088. doi:10.1126/science.1069268
Warren JJ, Moore PB (2001) Application of dipolar coupling data to the refinement of the solution structure of the sarcin-ricin loop RNA. J Biomol NMR 20(4):311–323
Warren JJ, Moore PB (2001) A maximum likelihood method for determining D(a)(PQ) and R for sets of dipolar coupling data. J Magn Reson 149(2):271–275. doi:10.1006/jmre.2001.2307
Vallurupalli P, Moore PB (2003) The solution structure of the loop E region of the 5S rRNA from spinach chloroplasts. J Mol Biol 325(5):843–856
Lee JH, Canny MD, De Erkenez A, Krilleke D, Ng YS, Shima DT, Pardi A, Jucker F (2005) A therapeutic aptamer inhibits angiogenesis by specifically targeting the heparin binding domain of VEGF165. Proc Natl Acad Sci USA 102(52):18902–18907. doi:10.1073/pnas.0509069102
Lee JH, Jucker F, Pardi A (2008) Imino proton exchange rates imply an induced-fit binding mechanism for the VEGF165-targeting aptamer, Macugen. FEBS Lett 582(13):1835–1839. doi:10.1016/j.febslet.2008.05.003
Ruckman J, Green LS, Beeson J, Waugh S, Gillette WL, Henninger DD, Claesson-Welsh L, Janjic N (1998) 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J Biol Chem 273(32):20556–20567
Acknowledgments
The Faculty of Science at the University of Gothenburg and the Swedish research council (VR) are gratefully acknowledged for financial support.
Author information
Authors and Affiliations
Corresponding author
Additional information
This paper belongs to Topical Collection 9th European Conference on Computational Chemistry (EuCo-CC9)
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 168 kb)
Rights and permissions
About this article
Cite this article
Eriksson, E.S.E., Joshi, L., Billeter, M. et al. De novo tertiary structure prediction using RNA123—benchmarking and application to Macugen. J Mol Model 20, 2389 (2014). https://doi.org/10.1007/s00894-014-2389-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-014-2389-z