Abstract
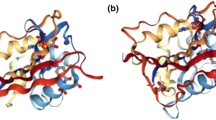
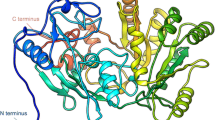
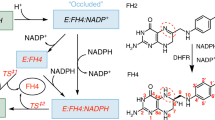
Human dihydrofolate reductase-like 1 (DHFRL1) has been identified as a second human dihydrofolate reductase (DHFR) enzyme. Although DHFRL1 have high sequence homology with human DHFR, dihydrofolate (DHF) exhibits a lowered binding affinity to DHFRL1 and the corresponding molecular mechanism is still unknown. To address this question, we studied the binding of DHF to DHFRL1 and DHFR by using molecular dynamics simulation. Moreover, to investigate the role the 24th residue of DHFR/DHFRL1 plays in DHF binding, R24W DHFRL1 mutant was also studied. The van der Waals interaction are more crucial for the total DHF binding energies, while the difference between the DHF binding energies of human DHFR and DHFRL1 can be attributed to the electrostatic interaction and the polar desolvation free energy. More specifically, lower DHF affinity to DHFRL1 can be mainly attributed to the reduction of net electrostatic interactions of residues Arg32 and Gln35 of DHFRL1 with DHF as being affected by Arg24. The side chain of Arg24 in DHFRL1 can extend deeply into the binding sites of DHF and NADPH, and disturb the DHF binding by steric effect, which rarely happens in human DHFR and R24W DHFRL1 mutant. Additionally, the conformation of loop I in DHFRL1 was also studied in this work. Interestingly, the loop conformation resemble to normal closed state of Escherichia coli DHFR other than the closed state of human DHFR. We hope this work will be useful to understand the general characteristics of DHFRL1.







Similar content being viewed by others
References
Roth B, Aig E, Rauckman BS, Strelitz JZ, Phillips AP, Ferone R, Bushby SR, Sigel CW (1981) 2,4-Diamino-5-benzylpyrimidines and analogues as antibacterial agents. 5. 3',5'-Dimethoxy-4'-substituted-benzyl analogues of trimethoprim. J Med Chem 24:933–941
Bertino JR, Sobrero A, Mini E, Moroson BA, Cashmore A (1987) Design and rationale for novel antifolates. NCI Monogr 5:87–91
Plowe CV, Kublin PG, Dzinjalamala FK, Kamwendo DS, Mukadam RAG, Chimpeni P, Molyneux ME, Taylor TE (2004) Sustained clinical efficacy of sulfadoxine-pyrimethamine for uncomplicated falciparum malaria in Malawi after 10 years as first line treatment: five year prospective study. Brit Med J 328:545–548
Fleming GF, Schilsky RL (1992) Antifolates: the next generation. Semin Oncol 19:707–719
Bertino JR (2009) Cancer research: from folate antagonism to molecular targets. Best Pract Res Clin Haematol 22:577–582
Anderson AC (2005) Targeting DHFR in parasitic protozoa. Drug Discov Today 10:121–128
Benkovic SJ (1980) On the mechanism of action of folate- and biopterin-requiring enzymes. Annu Rev Biochem 49:227–251
Maurer BJ, Barker PE, Masters JN, Ruddle FH, Attardi G (1984) Human dihydrofolate reductase gene is located in chromosome 5 and is unlinked to the related pseudogenes. Proc Natl Acad Sci U S A 81:1484–1488
McEntee G, Minguzzi S, O'Brien K, Ben Larbi N, Loscher C, O'Fagain C, Parle-McDermott A (2011) The former annotated human pseudogene dihydrofolate reductase-like 1 (DHFRL1) is expressed and functional. Proc Natl Acad Sci U S A 108:15157–15162
Anderson DD, Quintero CM, Stover PJ (2011) Identification of a de novo thymidylate biosynthesis pathway in mammalian mitochondria. Proc Natl Acad Sci U S A 108:15163–15168
Chunduru SK, Cody V, Luft JR, Pangborn W, Appleman JR, Blakley RL (1994) Methotrexate-resistant variants of human dihydrofolate reductase. Effects of Phe31 substitutions. J Biol Chem 269:9547–9555
Volpato JP, Yachnin BJ, Blanchet J, Guerrero V, Poulin L, Fossati E, Berghuis AM, Pelletier JN (2009) Multiple conformers in active site of human dihydrofolate reductase F31R/Q35E double mutant suggest structural basis for methotrexate resistance. J Biol Chem 284:20079–20089
Lewis WS, Cody V, Galitsky N, Luft JR, Pangborn W, Chunduru SK, Spencer HT, Appleman JR, Blakley RL (1995) Methotrexate-resistant variants of human dihydrofolate reductase with substitutions of leucine 22. Kinetics, crystallography, and potential as selectable markers. J Biol Chem 270:5057–5064
Rastelli G, Rio AD, Degliesposti G, Sgobba M (2010) Fast and accurate predictions of binding free energies using MM-PBSA and MM-GBSA. J Comput Chem 31:797–810
Cummins PL, Gready JE (2000) QM/MM and SCRF studies of the ionization state of 8-methylpterin substrate bound to dihydrofolate reductase: existence of a low-barrier hydrogen bond. J Mol Graph Model 18:42–49
Tosso RD, Andujar SA, Gutierrez L, Angelina E, Rodríguez R, Nogueras M, Baldoni H, Suvire FD, Cobo J, Enriz RD (2013) Molecular modeling study of dihydrofolate reductase inhibitors. molecular dynamics simulations, quantum mechanical calculations, and experimental corroboration. J Chem Inf Model. doi:10.1021/ci400178h
Oliveira AA, Rennó MN, de Matos CAS, Bertuzzi MD, Ramalho TC, Fraga CAM, França TCC (2011) Molecular modeling studies of Yersinia pestis dihydrofolate reductase. J Biomol Struct Dyn 29:351–367
Gorse AD, Gready JE (1997) Molecular dynamics simulations of the docking of substituted N5-deazapterins to dihydrofolate reductase. Protein Eng 10:23–30
Choowongkomon K, Theppabutr S, Songtawee N, Day N, White N, Woodrow C, Imwong M (2010) Computational analysis of binding between malarial dihydrofolate reductases and anti-folates. Malar J 9:65
Fan Y, Cembran A, Ma S, Gao J (2013) Connecting protein conformational dynamics with catalytic function as illustrated in dihydrofolate reductase. Biochemistry 52:2036–2049
Schnell JR, Dyson HJ, Wright PE (2004) Structure, dynamics, and catalytic function of dihydrofolate reductase. Annu Rev Biophys Biomol Struct 33:119–140
Swanwick RS, Shrimpton PJ, Allemann RK (2004) Pivotal role of Gly 121 in dihydrofolate reductase from Escherichia coli: the altered structure of a mutant enzyme may form the basis of its diminished catalytic performance. Biochemistry 43:4119–4127
Beard WA, Appleman JR, Huang S, Delcamp TJ, Freisheim JH, Blakley RL (1991) Role of the conserved active site residue tryptophan-24 of human dihydrofolate reductase as revealed by mutagenesis. Biochemistry 30:1432–1440
Thillet J, Absil J, Stone SR, Pictet R (1988) Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate and trimethoprim. J Biol Chem 263:12500–12508
Wang W, Kollman PA (2000) Free energy calculations on dimer stability of the HIV protease using molecular dynamics and a continuum solvent model. J Mol Biol 303:567–582
Kuhn B, Kollman PA (2000) Binding of a diverse set of ligands to Avidin and Streptavidin: an accurate quantitative prediction of their relative affinities by a combination of molecular mechanics and continuum solvent models. J Med Chem 43:3786–3791
Hou T, Zhu L, Chen L, Xu X (2002) Mapping the binding site of a large set of quinazoline type EGF-R inhibitors using molecular field analyses and molecular docking studies. J Chem Inf Comput Sci 43:273–287
Gohlke H, Case DA (2004) Converging free energy estimates: MM-PB(GB)SA studies on the protein–protein complex Ras–Raf. J Comput Chem 25:238–250
Hou T, Zhang W, Case DA, Wang W (2008) Characterization of domain-peptide interaction interface: a case study on the amphiphysin-1 SH3 domain. J Mol Biol 376:1201–1214
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucleic Acids Res 28:235–242
SYBYL molecular simulation package. 2004, http://www.sybyl.com.
Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ (2005) The Amber biomolecular simulation programs. J Comput Chem 26:1668–1688
Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P (2003) A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem 24:1999–2012
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general amber force field. J Comput Chem 25:1157–1174
Frisch MJ TG, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nak-ajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Laham A, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA (2003) Gaussian 03, revision C.02. Wallingford
Bayly CI, Cieplak P, Cornell W, Kollman PA (1993) A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J Phys Chem 97:10269–10280
Wang J, Wang W, Kollman PA, Case DA (2006) Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graph Model 25:247–260
Cummins PL, Ramnarayan K, Singh UC, Gready JE (1991) Molecular dynamics/free energy perturbation study on the relative affinities of the binding of reduced and oxidized NADP to dihydrofolate reductase. J Am Chem Soc 113:8247–8256
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935
Ryckaert J-P, Ciccotti G, Berendsen HJC (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23:327–341
Essmann U, Perera L, Berkowitz M, Darden T, Lee H, Pedersen L (1995) A smooth particle mesh Ewald method. J Chem Phys 103:8577–8593
Wang J, Hou T, Xu X (2006) Recent advances in free energy calculations with a combination of molecular mechanics and continuum models. Curr Comput Aided Drug Des 2:287–306
Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE (2000) Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res 33:889–897
Onufriev A, Bashford D, Case DA (2004) Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins 55:383–394
Weiser J, Shenkin PS, Still WC (1999) Approximate atomic surfaces from linear combinations of pairwise overlaps (LCPO). J Comput Chem 20:217–230
Hou T, Wang J, Li Y, Wang W (2010) Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model 51:69–82
Lee J, Kim J-S, Seok C (2010) Cooperativity and specificity of Cys2His2 zinc finger protein-DNA interactions: a molecular dynamics simulation study. J Phys Chem B 114:7662–7671
Gohlke H, Kiel C, Case DA (2003) Insights into protein-protein binding by binding free energy calculation and free energy decomposition for the Ras-Raf and Ras-RalGDS complexes. J Mol Biol 330:891–913
Bag S, Tawari NR, Degani MS, Queener SF (2010) Design, synthesis, biological evaluation and computational investigation of novel inhibitors of dihydrofolate reductase of opportunistic pathogens. Bioorg Med Chem 18:3187–3197
Sawaya MR, Kraut J (1997) Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: crystallographic evidence. Biochemistry 36:586–603
Davies JF, Delcamp TJ, Prendergast NJ, Ashford VA, Freisheim JH, Kraut J (1990) Crystal structures of recombinant human dihydrofolate reductase complexed with folate and 5-deazafolate. Biochemistry 29:9467–9479
Oefner C, D'Arcy A, Winkler FK (1988) Crystal structure of human dihydrofolate reductase complexed with folate. Eur J Biochem 174:377–385
Acknowledgments
We thank Prof. Jingyuan Li for helpful discussions. This work was supported by the Natural Science Foundation of China (No. 21173264) and the Foundation of Knowledge Innovative Engineering of Chinese Academy of Sciences (No. ZNWH-2011-011).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 2529 kb)
Rights and permissions
About this article
Cite this article
Gao, J., Cui, W., Du, Y. et al. Insight into the molecular mechanism about lowered dihydrofolate binding affinity to dihydrofolate reductase-like 1 (DHFRL1). J Mol Model 19, 5187–5198 (2013). https://doi.org/10.1007/s00894-013-2018-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-013-2018-2