Abstract
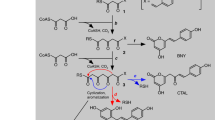
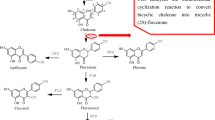
Chalcone isomerase (CHI) catalyzes the intramolecular cyclization of chalcones into flavonoids. The activity of CHI is essential for the biosynthesis of flavonoids precursors of floral pigments and phenylpropanoid plant defense compounds. In the present study, we explored the detailed binding structures and binding free energies for two different active site conformations of CHI with s-cis/s-trans conformers of three chalcone compounds by performing molecular dynamics (MD) simulations and binding free energy calculations. The computational results indicate that s-cis/s-trans conformers of chalcone compounds are orientated in the similar binding position in the active site of CHI and stabilized by the different first hydrogen bond network and the same second hydrogen bond network. The first hydrogen bond network results in much lower binding affinity of s-trans conformer of chalcone compound with CHI than that of s-cis conformer. The conformational change of the active site residue T48 from indirectly interacting with the substrate via the second hydrogen bond network to directly forming the hydrogen bond with the substrates cannot affect the binding mode of both conformers of chalcone compounds, but remarkably improves the binding affinity. These results show that CHI has a strong stereoselectivity. The calculated binding free energies for three chalcone compounds with CHI are consistent with the experimental activity data. In addition, several valuable insights are suggested for future rational design and discovery of high-efficiency mutants of CHI.

Stereoselectivity of chalcone isomerase with chalcone derivatives






Similar content being viewed by others
References
Dixon RA, Paiva NL (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7(7):1085–1097
Dooner HK, Robbins TP, Jorgensen RA (1991) Genetic and developmental control of anthocyanin biosynthesis. Annu Rev Genet 25:173–199
Long SR (1989) Rhizobium-legume nodulation: life together in the underground. Cell 56(2):203–214
Dixon RA, Steele CL (1999) Flavonoids and isoflavonoids - a gold mine for metabolic engineering. Trends Plant Sci 4(10):394–400
Setchell KD, Cassidy A (1999) Dietary isoflavones: biological effects and relevance to human health. J Nutr 129(3):758S–767S
Bednar RA, Hadcock JR (1988) Purification and characterization of chalcone isomerase from soybeans. J Biol Chem 263(20):9582–9588
Jung W, Yu O, Lau SM, O’Keefe DP, Odell J, Fader G, McGonigle B (2000) Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat Biotechnol 18(2):208–212
DellaPenna D (1999) Nutritional genomics: manipulating plant micronutrients to improve human health. Science 285(5426):375–379
Jez JM, Bowman ME, Dixon RA, Noel JP (2000) Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase. Nat Struct Biol 7(9):786–791
Jez JM, Bowman ME, Noel JP (2002) Role of hydrogen bonds in the reaction mechanism of chalcone isomerase. Biochemistry 41(16):5168–5176
Jez JM, Noel JP (2002) Reaction mechanism of chalcone isomerase. pH dependence, diffusion control, and product binding differences. J Biol Chem 277(2):1361–1369
Ngaki MN, Louie GV, Philippe RN, Manning G, Pojer F, Bowman ME, Li L, Larsen E, Wurtele ES, Noel JP (2012) Evolution of the chalcone-isomerase fold from fatty-acid binding to stereospecific catalysis. Nature 485(7399):530–533
Ruiz-Pernía JJ, Silla E, Tuñón I (2007) Enzymatic effects on reactant and transition states. The case of chalcone Isomerase. J Am Chem Soc 129(29):9117–9124
Hur S, Newby ZER, Bruice TC (2004) Transition state stabilization by general acid catalysis, water expulsion, and enzyme reorganization in Medicago savita chalcone isomerase. Proc Natl Acad Sci 101(9):2730–2735
Ruiz-Pernía JJ, Silla E, Tuñón I (2006) Comparative computational analysis of different active site conformations and substrates in a chalcone isomerase catalyzed reaction. J Phys Chem B 110(41):20686–20692
Ruiz-Pernía JJ, Ia T, Moliner V, Hynes JT, Roca M (2008) Dynamic effects on reaction rates in a Michael addition catalyzed by chalcone isomerase. Beyond the frozen environment approach. J Am Chem Soc 130(23):7477–7488
Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ (2005) The Amber biomolecular simulation programs. J Comput Chem 26(16):1668–1688
Case DAD, T.A., Cheatham TE, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, Walker RC, Zhang W, Wang B, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Schafmeister C, Ross WS, Kollman PA (2006). Amber 9, University of California: San Francisco
Frisch MJT, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JJA, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA (2004) Gaussian 03, Revision E01. Gaussian Inc, Wallingford
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79(2):926–935
Miyamoto S, Kollman PA (1992) Settle - an analytical version of the shake and rattle algorithm for rigid water models. J Comput Chem 13(8):952–962
Ryckaert JP, Ciccotti G, Berendsen HJC (1977) Numerical-integration of Cartesian equations of motion of a system with constraints - molecular-dynamics of N-alkanes. J Comput Phys 23(3):327–341
Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE (2000) Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res 33(12):889–897
Gilson MK, Sharp KA, Honig BH (1988) Calculating the electrostatic potential of molecules in solution: method and error assessment. J Comput Chem 9(4):327–335
Fu G, Liu H, Doerksen RJ (2012) Molecular modeling to provide insight into the substrate binding and catalytic mechanism of human biliverdin-IXα reductase. J Phys Chem B 116(32):9580–9594
Kuhn B, Kollman PA (2000) Binding of a diverse set of ligands to avidin and streptavidin: an accurate quantitative prediction of their relative affinities by a combination of molecular mechanics and continuum solvent models. J Med Chem 43(20):3786–3791
Villà J, Štrajbl M, Glennon TM, Sham YY, Chu ZT, Warshel A (2000) How important are entropic contributions to enzyme catalysis? Proc Natl Acad Sci 97(22):11899–11904
Zhang Y, Pan DB, Shen YL, Jin NZ, Liu HX, Yao XJ (2012) Understanding the molecular mechanism of the broad and potent neutralization of HIV-1 by antibody VRC01 from the perspective of molecular dynamics simulation and binding free energy calculations. J Mol Model 18(9):4517–4527
Acknowledgments
This work was supported by the Major State Basic Research Development Programs of China (2011CBA00701, 2012CB721003), the National Natural Science Foundation of China (20973049), the Doctoral Foundation by the Ministry of Education of China (20112303110005), the SF for leading experts in academe of Harbin of China (2011RFJGS026), the Science Funds for Distinguished Young Scholar of Heilongjiang Province (JC201206), and the Fundamental Research Funds for the Central Universities (Grant No. HIT. NSRIF. 2013056).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Yao, Y., Zhang, H. & Li, ZS. Stereoselectivity of chalcone isomerase with chalcone derivatives: a computational study. J Mol Model 19, 4753–4761 (2013). https://doi.org/10.1007/s00894-013-1975-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-013-1975-9