Abstract
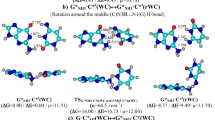
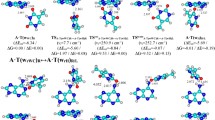
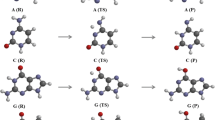
Combining quantum-mechanical (QM) calculations with quantum theory of atoms in molecules (QTAIM) and using the methodology of sweeps of the energetic, electron-topological, geometric and polar parameters, which describe the course of the tautomerization along the intrinsic reaction coordinate (IRC), we showed for the first time that the biologically important A∙A* base pair (Cs symmetry) formed by the amino and imino tautomers of adenine (A) tautomerizes via asynchronous concerted double proton transfer (DPT) through a transition state (TS), which is the A+∙A− zwitterion with the separated charge, with Cs symmetry. The nine key points, which can be considered as electron-topological “fingerprints” of the asynchronous concerted A∙A*↔A*∙A tautomerization process via the DPT, were detected and completely investigated along the IRC of the A∙A*↔A*∙A tautomerization. Based on the sweeps of the H-bond energies, it was found that intermolecular antiparallel N6Н⋯N6 (7.01 kcal mol−1) and N1H⋯N1 (6.88 kcal mol−1) H-bonds are significantly cooperative and mutually reinforce each other. It was shown for the first time that the A∙A*↔A*∙A tautomerization is assisted by the third C2H⋯HC2 dihydrogen bond (DHB), which, in contrast to the two others N6H⋯N6 and N1H⋯N1 H-bonds, exists within the IRC range from −2.92 to 2.92 Å. The DHB cooperatively strengthens, reaching its maximum energy 0.42 kcal mol−1 at IRC = −0.52 Å and minimum energy 0.25 kcal mol−1 at IRC = −2.92 Å, and is accompanied by strengthening of the two other aforementioned classical H-bonds. We established that the C2H⋯HC2 DHB completely satisfies the electron-topological criteria for H-bonding, in particular Bader’s and all eight “two-molecule” Koch and Popelier’s criteria. The positive value of the Grunenberg’s compliance constant (5.203 Å/mdyn) at the TSA∙A*↔A*∙A proves that the C2H⋯HC2 DHB is a stabilizing interaction. NBO analysis predicts transfer of charge from σ(C2–H) bonding orbital to σ*(H–C2) anti-bonding orbital; at this point, the stabilization energy E(2) is equal to 0.19 kcal mol−1 at the TSA∙A*↔A*∙A.









Similar content being viewed by others
References
Lehninger AL (1970) Biochemistry: the molecular basis of cell structure and function. Worth, New York
Rich A (1964) In: Tumerman LA (ed) Horizons in biochemistry. Mir, Moscow
Brovarets’ OO, Hovorun DM (2013) Can tautomerization of the A∙T Watson-Crick base pair via double proton transfer provoke point mutations during DNA replication? A comprehensive QM and QTAIM analysis. J Biomol Struct Dynam. doi:10.1080/07391102.2012.755795
Hovorun DM (1997) A structural isomerism of canonical nucleotide bases: AM1 calculation. Biopolym Cell 13:127–134
Samijlenko SP, Krechkivska OM, Kosach DA, Hovorun DM (2004) Transitions to high tautomeric states can be induced in adenine by interactions with carboxylate and sodium ions: DFT calculation data. J Mol Struct 708:97–104
Danilov VI, Anisimov VM, Kurita N, Hovorun D (2005) MP2 and DFT studies of the DNA rare base pairs: the molecular mechanism of the spontaneous substitution mutations conditioned by tautomerism of bases. Chem Phys Lett 412:285–293
Brovarets’ OO, Kolomiets’ IM, Hovorun DM (2012) Elementary molecular mechanisms of the spontaneous point mutations in DNA: a novel quantum-chemical insight into the classical understanding. In: Tada T (ed) Quantum chemistry—molecules for innovations. Rijeka, In Tech Open Access, pp 59–102
Fonseca Guerra C, Bickelhaupt FM, Saha S, Wang F (2006) Adenine tautomers: relative stabilities, ionization energies, and mismatch with cytosine. J Phys Chem A 110:4012–4020
Sabio M, Topiol S, Lumma WC (1990) An investigation of tautomerism in adenine and guanine. J Phys Chem 94:1366–1372
Kwiatkowski JS, Leszczynski J (1992) An ab initio quantum-mechanical study of tautomerism of purine, adenine and guanine. J Mol Struct (THEOCHEM) 208:35–44
Salter LM, Chaban GM (2002) Theoretical study of gas phase tautomerization reactions for the ground and first excited electronic states of adenine. J Phys Chem A 106:4251–4256
Singh RK, Ortiz JV, Mishra MK (2010) Tautomeric forms of adenine: vertical ionization energies and Dyson orbitals. Int J Quantum Chem 110:1901–1915
Trakhanov SD, Yusupov MM, Agalarov SC, Garber MB, Ryazantsev SN, Tischenko SV, Shirokov VA (1987) Crystallization of 70 S ribosomes and 30 S ribosomal subunits from Thermus thermophilus. FEBS Lett 220:319–322
Wimberly BT, Brodersen DE, Clemons WM Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V (2000) Structure of the 30 S ribosomal subunit. Nature 407:327–339
Lee JC, Gutell RR (2004) Diversity of base-pair conformations and their occurrence in rRNA structure and RNA structural motifs. J Mol Biol 344:1225–1249
von Böhlen K, Makowski I, Hansen HAS, Bartels H, Berkovitch-Yellin Z, Zaytzev-Bashan A, Meyer S, Paulke C, Franceschi F, Yonath A (1991) Characterization and preliminary attempts for derivatization of crystals of large ribosomal subunits from Haloarcula marismortui diffracting to 3 Å resolution. J Mol Biol 222:11–15
Kondratyuk IV, Samijlenko SP, Kolomiets’ IM, Hovorun DM (2000) Prototropic molecular–zwitterionic tautomerism of xanthine and hypoxanthine. J Mol Struct 523:109–118
Sun X, Lee JK (2010) The stability of DNA duplexes containing hypoxanthine (inosine): gas versus solution phase and biological implications. J Org Chem 75:1848–1854
O’Brien PJ, Ellenberger T (2004) The Escherichia coli 3-methyladenine DNA glycosylase AlkA has a remarkably versatile active site. J Biol Chem 279:26876–26884
Hill-Perkins M, Jones MD, Karran P (1986) Sitespecific mutagenesis in vivo by single methylated or deaminated purine bases. Mutat Res 162:153–163
Basílio Janke EM, Riechert-Krause F, Weisz K (2011) Low-temperature NMR studies on inosine wobble base pairs. J Phys Chem B 115:8569–8574
Brovarets’ OO, Hovorun DM (2012) Prototropic tautomerism and basic molecular principles of hypoxanthine mutagenicity: an exhaustive quantum-chemical analysis. J Biomol Struct Dynam. doi:10.1080/07391102.2012.715041
Brovarets’ OO, Zhurakivsky RO, Hovorun DM (2013) The physico-chemical “anatomy” of the tautomerization through the DPT of the biologically important pairs of hypoxanthine with DNA bases: QM and QTAIM perspectives. J Mol Model. doi:10.1007/s00894-012-1720-9
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Pople JA et al (2010) GAUSSIAN 09 (Revision B.01). Gaussian, Wallingford
Parr RG, Yang W (1989) Density-functional theory of atoms and molecules. Oxford University Press, Oxford
Tirado-Rives J, Jorgensen WL (2008) Performance of B3LYP density functional methods for a large set of organic molecules. J Chem Theory Comput 4:297–306
Becke AD (1988) Density-functional exchange-energy approximation with correct asymptotic behaviour. Phys Rev A 38:3098–3100
Lee C, Yang W, Parr RG (1988) Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B 37:785–789
Brovarets’ OO, Hovorun DM (2010) How stable are the mutagenic tautomers of DNA bases? Biopolym Cell 26:72–76
Brovarets’ OO, Hovorun DM (2010) Stability of mutagenic tautomers of uracil and its halogen derivatives: the results of quantum-mechanical investigation. Biopolym Cell 26:295–298
Brovarets’ OO, Hovorun DM (2010) Quantum-chemical investigation of tautomerization ways of Watson-Crick DNA base pair guanine-cytosine. Ukr Biochem J 82:55–60
Brovarets’ OO, Hovorun DM (2010) Molecular mechanisms of transitions induced by cytosine analogue: comparative quantum-chemical study. Ukr Biochem J 82:51–56
Brovarets’ OO, Hovorun DM (2010) Quantum-chemical investigation of the elementary molecular mechanisms of pyrimidine-purine transversions. Ukr Biochem J 82:57–67
Brovarets’ OO, Zhurakivsky RO, Hovorun DM (2010) Is there adequate ionization mechanism of the spontaneous transitions? Quantum-chemical investigation. Biopolym Cell 26:398–405
Brovarets’ OO, Hovorun DM (2011) IR Vibrational spectra of H-bonded complexes of adenine, 2-aminopurine and 2-aminopurine+ with cytosine and thymine: quantum-chemical study. Opt Spectrosc 111:750–757
Brovarets’ OO, Hovorun DM (2011) Intramolecular tautomerization and the conformational variability of some classical mutagens—cytosine derivatives: quantum chemical study. Biopolym Cell 27:221–230
Frisch MJ, Head-Gordon M, Pople JA (1990) Semi-direct algorithms for the MP2 energy and gradient. Chem Phys Lett 166:281–289
Frisch MJ, Pople JA, Binkley JS (1984) Self-consistent molecular orbital methods 25. Supplementary functions for Gaussian basis sets. J Chem Phys 80:3265–3269
Hariharan PC, Pople JA (1973) The influence of polarization functions on molecular orbital hydrogenation energies. Theor Chem Accounts Theor Comput Model 28:213–222
Krishnan R, Binkley JS, Seeger R, Pople JA (1980) Self-consistent molecular orbital methods. XX. A basis set for correlated wave functions. J Chem Phys 72:650–654
Dunning Jr TH (1989) Gaussian basis sets for use in correlated molecular calculations. I. The atoms boron through neon and hydrogen. J Chem Phys 90:1007–1023
Kendall RA, Dunning Jr TH, Harrison RJ (1992) Electron affinities of the first-row atoms revisited. Systematic basis sets and wave functions. J Chem Phys 96:6796–6806
Brovarets’ OO, Yurenko YP, Dubey IY, Hovorun DM (2012) Can DNA-binding proteins of replisome tautomerize nucleotide bases? Ab initio model study. J Biomol Struct Dynam 29:1101–1109
Pelmenschikov A, Hovorun DM, Shishkin OV, Leszczynski J (2000) A density functional theory study of vibrational coupling between ribose and base rings of nucleic acids with ribosyl guanosine as a model system. J Chem Phys 113:5986–5990
Shishkin OV, Pelmenschikov A, Hovorun DM, Leszczynski J (2000) Theoretical analysis of low-lying vibrational modes of free canonical 2′-deoxyribonucleosides. Chem Phys 260:317–325
Platonov MO, Samijlenko SP, Sudakov OO, Kondratyuk IV, Hovorun DM (2005) To what extent can methyl derivatives be regarded as stabilized tautomers of xanthine? Spectrochim Acta A Mol Biomol Spectrosc 62:112–114
Danilov VI, van Mourik T, Kurita N, Wakabayashi H, Tsukamoto T, Hovorun DM (2009) On the mechanism of the mutagenic action of 5-bromouracil: a DFT study of uracil and 5-bromouracil in a water cluster. J Phys Chem A 113:2233–2235
Yurenko YP, Zhurakivsky RO, Ghomi M, Samijlenko SP, Hovorun DM (2007) Comprehensive conformational analysis of the nucleoside analogue 2′-β-deoxy-6-azacytidine by DFT and MP2 calculations. J Phys Chem B 111:6263–6271
Yurenko YP, Zhurakivsky RO, Samijlenko SP, Ghomi M, Hovorun D (2007) The whole of intramolecular H-bonding in the isolated DNA nucleoside thymidine. AIM electron density topological study. Chem Phys Lett 447:140–146
Yurenko YP, Zhurakivsky RO, Ghomi M, Samijlenko SP, Hovorun DM (2007) How many conformers determine the thymidine low-temperature matrix infrared spectrum? DFT and MP2 quantum chemical study. J Phys Chem B 111:9655–9663
Yurenko YP, Zhurakivsky RO, Ghomi M, Samijlenko SP, Hovorun DM (2008) Ab initio comprehensive conformational analysis of 2′-deoxyuridine, the biologically significant DNA minor nucleoside, and reconstruction of its low-temperature matrix infrared spectrum. J Phys Chem B 112:1240–1250
Yurenko YP, Zhurakivsky RO, Samijlenko SP, Hovorun DM (2011) Intramolecular CH ⋯O hydrogen bonds in the AI and BI DNA-like conformers of canonical nucleosides and their Watson-Crick pairs. Quantum chemical and AIM analysis. J Biomol Struct Dynam 29:51–65
Ponomareva AG, Yurenko YP, Zhurakivsky RO, van Mourik T, Hovorun DM (2012) Complete conformational space of the potential HIV-1 reverse transcriptase inhibitors d4U and d4C. A quantum chemical study. Phys Chem Chem Phys 14:6787–6795
Matta CF (2010) How dependent are molecular and atomic properties on the electronic structure method? Comparison of Hartree-Fock, DFT, and MP2 on a biologically relevant set of molecules. J Comput Chem 31:1297–1311
Lozynski M, Rusinska-Roszak D, Mack H-G (1998) Hydrogen bonding and density functional calculations: the B3LYP approach as the shortest way to MP2 results. J Phys Chem A 102:2899–2903
Hovorun DM, Gorb L, Leszczynski J (1999) From the nonplanarity of the amino group to the structural nonrigidity of the molecule: a post-Hartree-Fock ab initio study of 2-aminoimidazole. Int J Quantum Chem 75:245–253
Peng C, Schlegel HB (1993) Combining synchronous transit and quasi-Newton methods to find transition states. Isr J Chem 33:449–454
Peng C, Ayala PY, Schlegel HB, Frisch MJ (1996) Using redundant internal coordinates to optimize equilibrium geometries and transition states. J Comput Chem 17:49–56
Hratchian HP, Schlegel HB (2004) Accurate reaction paths using a Hessian based predictor-corrector integrator. J Chem Phys 120:9918–9924
Hratchian HP, Schlegel HB (2005) Finding minima, transition states, and following reaction pathways on ab initio potential energy surfaces. In: Dykstra CE, Frenking G, Kim KS, Scuseria G (eds) Theory and applications of computational chemistry: the first 40 years. Elsevier, Amsterdam, pp 195–249
Hratchian HP, Schlegel HB (2005) Using Hessian updating to increase the efficiency of a Hessian based predictor-corrector reaction path following method. J Chem Theory Comput 1:61–69
Boys SF, Bernardi F (1970) The calculation of small molecular interactions by the differences of separate total energies. Some procedures with reduced errors. Mol Phys 19:553–566
Gutowski M, Van Lenthe JH, Verbeek J, Van Duijneveldt FB, Chalasinski G (1986) The basis set superposition error in correlated electronic structure calculations. Chem Phys Lett 124:370–375
Sordo JA, Chin S, Sordo TL (1988) On the counterpoise correction for the basis set superposition error in large systems. Theor Chim Acta 74:101–110
Sordo JA (2001) On the use of the Boys–Bernardi function counterpoise procedure to correct barrier heights for basis set superposition error. J Mol Struct (THEOCHEM) 537:245–251
Atkins PW (1998) Physical chemistry. Oxford University Press, Oxford
Wigner E (1932) Über das Überschreiten von potentialschwellen bei chemischen Reaktionen [crossing of potential thresholds in chemical reactions]. Zeit Physik Chem B19:203–216
Bader RFW (1990) Atoms in molecules: a quantum theory. Oxford University Press, Oxford
Keith TA (2011) AIMAll (Version 11.12.19) Retrieved from http://aim.tkgristmill.com
Koch U, Popelier PLA (1995) Characterization of C-H-O hydrogen bonds on the basis of the charge density. J Phys Chem 99:9747–9754
Iogansen AV (1999) Direct proportionality of the hydrogen bonding energy and the intensification of the stretching ν(XH) vibration in infrared spectra. Spectrochim Acta A Mol Biomol Spectrosc 55:1585–1612
Espinosa E, Molins E, Lecomte C (1998) Hydrogen bond strengths revealed by topological analyses of experimentally observed electron densities. Chem Phys Lett 285:170–173
Mata I, Alkorta I, Espinosa E, Molins E (2011) Relationships between interaction energy, intermolecular distance and electron density properties in hydrogen bonded complexes under external electric fields. Chem Phys Lett 507:185–189
Brandhorst K, Grunenberg J (2010) Efficient computation of compliance matrices in redundant internal coordinates from Cartesian Hessians for nonstationary points. J Chem Phys 132:184101–184107
Brandhorst K, Grunenberg J (2008) How strong is it? The interpretation of force and compliance constants as bond strength descriptors. Chem Soc Rev 37:1558–1567
Grunenberg J, Barone G (2013) Are compliance constants ill-defined descriptors for weak interactions? RSC Adv 3:4757–4762
Weinhold F, Landis C (2005) Valency and bonding. A natural bond orbital donor-acceptor perspective. Cambridge University Press, Cambridge
Saenger W (1984) Principles of nucleic acid structure. Springer, New York
Govorun DN, Danchuk VD, Mishchuk YaR, Kondratyuk IV, Radomsky NF, Zheltovsky NV (1992) AM1 calculation of the nucleic acid bases structure and vibrational spectra. J Mol Struct 267:99–103
Nikolaienko TY, Bulavin LA, Hovorun DM (2011) How flexible are DNA constituents? The quantum-mechanical study. J Biomol Struct Dynam 29:563–575
Gribov LA, Mushtakova SP (1999) Quantum chemistry: textbook (Kvantovaya Khimiya: Uchebnik) (in Russian). Gardariki, Moscow, pp 317–319
Toro-Labbé A, Gutiérrez-Oliva S, Concha MC, Murray JS, Politzer P (2004) Analysis of two intramolecular proton transfer processes in terms of the reaction force. J Chem Phys 121:4570–4576
Burda JV, Toro-Labbé A, Gutiérrez-Oliva S, Murray JS, Politzer P (2007) Reaction force decomposition of activation barriers to elucidate solvent effects. J Phys Chem A 111:2455–2457
Jaque P, Toro-Labbé A, Politzer P, Geerlings P (2008) Reaction force constant and projected force constants of vibrational modes along the path of an intramolecular proton transfer reaction. Chem Phys Lett 456:135–140
Murray JS, Toro-Labbé A, Clark T, Politzer P (2009) Analysis of diatomic bond dissociation and formation in terms of the reaction force and the position-dependent reaction force constant. J Mol Model 15:701–706
Toro-Labbé A, Gutiérrez-Oliva S, Murray JS, Politzer P (2009) The reaction force and the transition region of a reaction. J Mol Model 15:707–710
Murray JS, Toro-Labbé A, Gutiérrez-Oliva S, Politzer P (2010) Identification of pseudodiatomic behavior in polyatomic bond dissociation: reaction force analysis. J Chem Phys 132:154308
Lipkowski P, Grabowski SJ, Robinson TL, Leszczynski J (2004) Properties of the C-H⋯H dihydrogen bond: an ab initio and topological analysis. J Phys Chem A 108:10865–10872
Grabowski SJ, Sokalski WA, Leszczynski (2005) How short can the H⋯H intermolecular contact be? New findings that reveal the covalent nature of extremely strong interactions. J Phys Chem A 109:4331–4341
Kaplan I (2006) Intermolecular interactions: physical picture, computational methods and model potentials (Wiley Series in Theoretical Chemistry). Wiley, Chichester
Mishchuk YaR, Potyagaylo AL, Hovorun DM (2000) Structure and dynamics of 6-azacytidine by MNDO/H quantum-chemical method. J Mol Struct 552:283–289
Ponomareva AG, Yurenko YP, Zhurakivsky RO, van Mourik T, Hovorun DM (2013) Structural and energetic properties of the potential HIV-1 reverse transcriptase inhibitors d4A and d4G: a comprehensive theoretical investigation. J Biomol Struct Dynam. doi:10.1080/07391102.2013.789401
Brovarets’ OO, Yurenko YP, Hovorun DM (2013) Intermolecular СН⋯О/N Н-bonds in the biologically important pairs of natural nucleobases: a thorough quantum-chemical study. J Biomol Struct Dynam. doi:10.1080/07391102.2013.799439
Bondi AJ (1964) Van der Waals volumes and radii. J Phys Chem 68:441–451
Gilli P, Bertolasi V, Ferretti V, Gilli G (1994) Evidence for resonance-assisted hydrogen bonding. 4. Covalent nature of the strong homonuclear hydrogen bond. Study of the O-H…O system by crystal structure correlation methods. J Am Chem Soc 116:909–915
Desiraju GR, Steiner T (1999) The weak hydrogen bond in structural chemistry and biology. Oxford University Press, New York
Lakshmi B, Samuelson AG, Jovan Jose KV, Gadre SR, Arunan E (2005) Is there a hydrogen bond radius? Evidence from microwave spectroscopy, neutron scattering and X-ray diffraction results. New J Chem 29:371–377
Acknowledgments
This work was partially supported by the Science and Technology Center in Ukraine (STCU) within the project № 5728 for years 2012–2014. O.O.B. was supported by a Grant of the President of Ukraine to support scientific research of young scientists for 2012 year from the State Fund for Fundamental Research of Ukraine (project № GP/F44/086) and by a Grant of the President of Ukraine for talented youth for year 2012 from the Ministry of Education and Science, Youth and Sports of Ukraine. The authors thank the Bogolyubov Institute for Theoretical Physics of the National Academy of Sciences of Ukraine for providing calculation resources and software. This work was performed using computational facilities of joint computer cluster of SSI “Institute for Single Crystals” of the National Academy of Sciences of Ukraine and Institute for Scintillation Materials of the National Academy of Sciences of Ukraine incorporated into Ukrainian National Grid.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 412 kb)
Rights and permissions
About this article
Cite this article
Brovarets’, O.O., Zhurakivsky, R.O. & Hovorun, D.M. DPT tautomerization of the long A∙A* Watson-Crick base pair formed by the amino and imino tautomers of adenine: combined QM and QTAIM investigation. J Mol Model 19, 4223–4237 (2013). https://doi.org/10.1007/s00894-013-1880-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-013-1880-2