Abstract
Survivin, the smallest inhibitor of apoptosis protein (IAP), is a valid target for cancer research. It mediates both the apoptosis pathway and the cell cycle and has been proposed to form a complex with the cyclin-dependent kinase protein CDK4. The resulting complex transports CDK4 from the cytosol to the nucleus, where CDK4 participates in cell division. Survivin has been recognized as a node protein that interacts with several partners; disruption of the formed complexes can lead to new anticancer compounds. We propose a rational model of the survivin/CDK4 complex that fulfills the experimental evidence and that can be used for structure-based design of inhibitors modifying its interface recognition. In particular, the suggested complex involves the alpha helical domain of survivin and resembles the mode of binding of survivin in the survivin/borealin X-ray structure. The proposed model has been obtained by combining protein–protein docking, fractal-based shape complementarity, electrostatics studies and extensive molecular dynamics simulations.

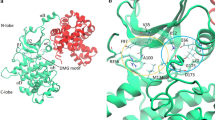
Proposed model of the survivin/CDK4 complex with a close view of the best model refined through molecular dynamics simulations





Similar content being viewed by others
References
Velculescu VE, Madden SL, Zhang L, Lash AE, Yu J, Rago C, Lal A, Wang CJ, Beaudry GA, Ciriello KM, Cook BP, Dufault MR, Ferguson AT, Gao Y, He TC, Hermeking H, Hiraldo SK, Hwang PM, Lopez MA, Luderer HF, Mathews B, Petroziello JM, Polyak K, Zawel L, Zhang W, Zhang X, Zhou W, Haluska FG, Jen J, Sukumar S, Landes GM, Riggins GJ, Vogelstein B, Kinzler KW (1999) Analysis of human transcriptomes. Nat Genet 23:387–388
Altieri DC (2008) Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer 8:61–70
Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N (2004) A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 351:2817–2826
Fesik SW (2005) Promoting apoptosis as a strategy for cancer drug discovery. Nat Rev Cancer 5:876–885
Sun C, Nettesheim D, Liu Z, Olejniczak ET (2005) Solution structure of human Survivin and its binding interface with Smac/Diablo. Biochemistry 44:11–17
Dohi T, Okada K, Xia F, Wilford CE, Samuel T, Welsh K, Marusawa H, Zou H, Armstrong R, Matsuzawa S, Salvesen GS, Reed JC, Altieri DC (2004) An IAP–IAP complex inhibits apoptosis. J Biol Chem 279:34087–34090
Fortugno P, Beltrami E, Plescia J, Fontana J, Pradhan D, Marchisio PC, Sessa WC, Altieri DC (2003) Regulation of survivin function by Hsp90. Proc Natl Acad Sci USA 100:13791–13796
Wheatley SP, Henzing AJ, Dodson H, Khaled W, Earnshaw WC (2004) Aurora-B phosphorylation in vitro identifies a residue of survivin that is essential for its localization and binding to inner centromere protein (INCENP) in vivo. J Biol Chem 279:5655–5660
Bourhis E, Hymowitz SG, Cochran AG (2007) The mitotic regulator Survivin binds as a monomer to its functional interactor Borealin. J Biol Chem 282:35018–35023
Suzuki A, Hayashida M, Ito T, Kawano H, Nakano T, Miura M, Akahane K, Shiraki K (2000) Survivin initiates cell cycle entry by the competitive interaction with Cdk4/p16INK4a and Cdk2/Cyclin E complex activation. Oncogene 19:3225–3234
Suzuki A, Ito T, Kawano H, Hayashida M, Hayasaki Y, Tsutomi Y, Akahane K, Nakano T, Miura M, Shiraki K (2000) Survivin initiates procaspase 3/p21 complex formation as a result of interaction with Cdk4 to resist Fas-mediated cell death. Oncogene 19:1346–1353
Ai MD, Li LL, Zhao XR, Wu Y, Gong JP, Cao Y (2005) Regulation of Survivin and CDK4 by Epstein-Barr virus encoded latent membrane protein 1 in nasopharyngeal carcinoma cell lines. Cell Res 15:777–784
Zhang L, Liu J, Lin H, Hu Q, Liu A, Hu Y (2006) Expression of survivin, CDK4, Ki-67 and clinical significance in pediatric acute leukemia. J Huazhong Univ Sci Technol 26:552–554
Chantalat L, Skoufias DA, Kleman JP, Jung B, Dideberg O, Margolis RL (2000) Crystal structure of human survivin reveals a bow tie-shaped dimer with two unusual alpha-helical extensions. Mol Cell 6:183–189
Day PJ, Cleasby A, Tickle IJ, O'Reilly M, Coyle JE, Holding FP, McMenamin RL, Yon J, Chopra R, Lengauer C, Jhoti H (2009) Crystal structure of human CDK4 in complex with a D-type cyclin. Proc Natl Acad Sci USA 106:4166–4170
Russo AA, Tong L, Lee JO, Jeffrey PD, Pavletich NP (1998) Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature 395:237–243
Humphrey W, Dalke A, Schulten K (1996) VMD-visual molecular dynamics. J Mol Graph 14:33–38
Eswar N, Mari-Renom MA, Webb B, Madhusudhan MS, Eramian D, Shen M, Pieper U, Sali A (2006) Comparative protein structure modeling with MODELLER. Current protocols in Bioinformatics. Supplement 15, Wiley, New York, pp 5.6.1–5.6.30, 200
MOE: Molecular Operating Environment; Chemical Computing Group, Inc. http://www.chemcomp.com/
Wang J, Cieplak P, Kollman PA (2000) How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J Comput Chem 21:1049–1074
Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ (2005) PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res 33:W363–W367
Gray JJ, Moughan SE, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, Baker D (2003) Protein–protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol 331:281–299
Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA 98:10037–10041
PyMOL Molecular Graphics System, Version 1.3, Schrödinger, LLC. http://www.pymol.org
Renthal R (1999) Transmembrane and water-soluble helix bundles display reverse patterns of surface roughness. Biochem Biophys Res Commun 263:714–717
Sanner MF, Olson AJ, Spehner JC (1996) Reduced surface: an efficient way to compute molecular surfaces. Biopolymers 38:305–320
Pang YP, Xu K, El Yazla J, Prendergast F (2000) Successful molecular dynamics simulation of the zinc-bound farnesyltransferase using the cationic dummy atom approach. Protein Sci 9:1857–1865
Obiol-Pardo C, Rubio-Martínez J (2007) Comparative evaluation of MMPBSA and XSCORE to compute binding free energy in XIAP-peptide complexes. J Chem Inf Model 47:134–142
Obiol-Pardo C, Granadino-Roldán JM, Rubio-Martínez J (2008) Protein–protein recognition as a first step towards the inhibition of XIAP and Survivin anti-apoptotic proteins. J Mol Recognit 21:190–204
Case DA, Darden TA, Cheatham TE III, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, Merz KM, Wang B, Hayik S, Roitberg A, Seabra G, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Mathews DH, Seetin MG, Sagui C, Babin V, Kollman PA (2008) AMBER 10. University of California, San Francisco
Harvey MJ, Giupponi G, de Fabritiis G (2009) ACEMD: accelerating biomolecular dynamics in the microsecond time scale. J Chem Theory Comput 5:1632–1639
Reynolds C, Damerell D, Jones S (2008) ProtorP: a protein—protein interaction analysis server. Bioinformatics 25:413–414
Nooren IM, Thornton JM (2003) Structural characterisation and functional significance of transient protein-protein interactions. J Mol Biol 325:991–1018
Kaczor AA, Guixà-González R, Carrió P, Obiol-Pardo C, Pastor M, Selent J (2012) Fractal dimension as a measure of surface roughness of G protein-coupled receptors: implications for structure and function. J Mol Model 18:4465–4467
Ross NT, Katt WP, Hamilton AD (2010) Synthetic mimetics of protein secondary structure domains. Phil Trans R Soc A 368:989–1008
Pavlyukov MS, Antipova NV, Balashova MV, Vinogradova TV, Kopantzev EP, Shakhparanov MI (2011) Survivin monomer plays an essential role in apoptosis regulation. J Biol Chem 286:23296–23307
Acknowledgments
This paper was funded by Foundation for Polish Science (FNP, Outgoing Fellowship Kolumb for Agnieszka A. Kaczor).
Author information
Authors and Affiliations
Corresponding author
Additional information
Jana Selent and Agnieszka A. Kaczor contributed equally to this work.
Rights and permissions
About this article
Cite this article
Selent, J., Kaczor, A.A., Guixà-González, R. et al. Rational design of the survivin/CDK4 complex by combining protein–protein docking and molecular dynamics simulations. J Mol Model 19, 1507–1514 (2013). https://doi.org/10.1007/s00894-012-1705-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-012-1705-8