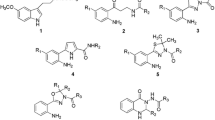
Abstract
Selective inhibition of the nitric oxide synthase isoforms (NOS) is a promising approach for the treatment of various disorders. However, given the high active site conservation among all NOS isoforms, the design of selective inhibitors is a challenging task. Analysis of the X-ray crystal structures of the NOS isoforms complexed with known inhibitors most often gives no clues about the structural determinants behind the selective inhibition since the inhibitors share the same binding conformation. Aimed at a better understanding of the structural factors responsible for selective inhibition of NOS isoforms we have performed MD simulations for iNOS, nNOS and eNOS complexed with Nω-NO2-L-Arg (1), and with the aminopyridine derivatives 2 and 3. The slightly better selectivity of 1 for nNOS may be assigned to the presence of extra charge–charge interactions due to its “extended” conformation. While the high affinity of 2 for iNOS can be explained by the formation of an iNOS-specific subpocket upon binding, the lack of affinity for eNOS is associated to a conformational change in Glu363. The strong van der Waals and electrostatic interactions between 3 and the active site of nNOS are most likely responsible for its higher affinity for this isoform. Owing to the elongated and narrow binding pocket of iNOS, the correct positioning of 3 over the heme group is difficult, which may account for its lower affinity toward this isoform. Brought together, our results might help to rationalize the design of selective NOS inhibitors.

Overall RMSD of the protein backbone over 8 ns simulation is shown for the complexes 3:eNOSmonomer and 3:eNOSdimer










Similar content being viewed by others
References
Alderton WK, Cooper CE, Knowles RG (2001) Nitric oxide synthases: structure, function and inhibition. Biochem J 357:593–615
Moncada S, Palmer RM, Higgs EA (1991) Nitric oxide: physiology, pathophysiology, and pharmacology. Pharmacol Rev 43(2):109–142
Li H, Poulos TL (2005) Structure-function studies on nitric oxide synthases. J Inorg Biochem 99(1):293–305
Leiper J, Nandi M (2011) The therapeutic potential of targeting endogenous inhibitors of nitric oxide synthesis. Nat Rev Drug Discov 10(4):277–291
Ji H, Erdal EP, Litzinger EA, Seo J, Zhu Y, Xue F, Fang J, Huang J, Silverman RB (2009) Selective neuronal nitric oxide synthase inhibitors. Front Med Chem 4:842–882
Wink DA, Mitchell JB (2003) Nitric oxide and cancer: an introduction. Free Radic Biol Med 34(8):951–954
Huang PL, Huang Z, Mashimo H, Bloch KD, Moskowitz MA, Bevan JA, Fishman MC (1995) Hypertension in mice lacking the gene for endothelial nitric oxide synthase. Nature 377(6546):239–242
Vallance P, Leiper J (2002) Blocking NO synthesis: how, where and why? Nat Rev Drug Discov 1(12):939–950
Di Giacomo C, Sorrenti V, Salerno L, Cardile V, Guerrera F, Siracusa MA, Avitabile M, Vanella A (2003) Novel inhibitors of neuronal nitric oxide synthase. Exp Biol Med 228(5):486–490
Zicha J, Pechánová O, Dobesová Z, Kunes J (2003) Hypertensive response to chronic NG-nitro-L-arginine methyl ester (L-NAME) treatment is similar in immature and adult Wistar rats. Clin Sci 105(4):483–489
Macdonald JE (1996) Chapter 23. Nitric oxide synthase inhibitors. Ann Rep Med Chem 31:221–230
Granik VG, Grigorev NB (2002) Nitric oxide synthase inhibitors: biology and chemistry. Russ Chem Bull 51(11):1973–1995
Tafi A, Angeli L, Venturini G, Travagli M, Corelli F, Botta M (2006) Computational studies of competitive inhibitors of nitric oxide synthase (NOS) enzymes: towards the development of powerful and isoform-selective inhibitors. Curr Med Chem 13(16):1929–1946
Oliveira BL, Correia JDG, Raposinho PD, Santos I, Ferreira A, Cordeiro C, Freire AP (2009) Re and 99mTc organometallic complexes containing pendant l-arginine derivatives as potential probes of inducible nitric oxide synthase. Dalton Trans 1:152–162
Oliveira BL, Raposinho PD, Mendes F, Figueira FV, Santos I, Ferreira AN, Cordeiro C, Freire AP, Correia JDG (2010) Re and Tc tricarbonyl complexes: from the suppression of NO biosynthesis in macrophages to in vivo targeting of inducible nitric oxide synthase. Bioconjug Chem 21(12):2168–2172
Oliveira BL, Raposinho PD, Mendes F, Santos IC, Santos I, Ferreira A, Cordeiro C, Freire AP, Correia JDG (2011) Targeting nitric oxide synthase with 99mTc/Re-tricarbonyl complexes containing pendant guanidino or isothiourea moieties. J Organomet Chem 696(5):1057–1065
Fischmann TO, Hruza A, Niu XD, Fossetta JD, Lunn CA, Dolphin E, Prongay AJ, Reichert P, Lundell DJ, Narula SK, Weber PC (1999) Structural characterization of nitric oxide synthase isoforms reveals striking active-site conservation. Nat Struct Mol Biol 6(3):233–242
Garvey EP, Oplinger JA, Furfine ES, Kiff RJ, Laszlo F, Whittle BJR, Knowles RG (1997) 1400W Is a slow, tight binding, and highly selective inhibitor of inducible nitric-oxide synthase in vitro and in vivo. J Biol Chem 272(8):4959–4963
Fedorov R, Hartmann E, Ghosh DK, Schlichting I (2003) Structural basis for the specificity of the nitric-oxide synthase inhibitors W1400 and Nω-Propyl-l-Arg for the inducible and neuronal isoforms. J Biol Chem 278(46):45818–45825
Li H, Raman CS, Martásek P, Masters BSS, Poulos TL (2001) Crystallographic studies on endothelial nitric oxide synthase complexed with nitric oxide and mechanism-based inhibitors†. Biochemistry 40(18):5399–5406
Ji H, Stanton BZ, Igarashi J, Li H, Martásek P, Roman LJ, Poulos TL, Silverman RB (2008) Minimal pharmacophoric elements and fragment hopping, an approach directed at molecular diversity and isozyme selectivity. Design of selective neuronal nitric oxide synthase inhibitors. J Am Chem Soc 130(12):3900–3914
Garcin ED, Arvai AS, Rosenfeld RJ, Kroeger MD, Crane BR, Andersson G, Andrews G, Hamley PJ, Mallinder PR, Nicholls DJ, St-Gallay SA, Tinker AC, Gensmantel NP, Mete A, Cheshire DR, Connolly S, Stuehr DJ, Aberg A, Wallace AV, Tainer JA, Getzoff ED (2008) Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase. Nat Chem Biol 4(11):700–707
Matter H, Kumar HSA, Fedorov R, Frey A, Kotsonis P, Hartmann E, Fröhlich LG, Reif A, Pfleiderer W, Scheurer P, Ghosh DK, Schlichting I, Schmidt HHHW (2005) Structural analysis of isoform-specific inhibitors targeting the tetrahydrobiopterin binding site of human nitric oxide synthases. J Med Chem 48(15):4783–4792
Li H, Raman CS, Martásek P, Král V, Masters BSS, Poulos TL (2000) Mapping the active site polarity in structures of endothelial nitric oxide synthase heme domain complexed with isothioureas. J Inorg Biochem 81(3):133–139
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19(14):1639–1662
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30(16):2785–2791
Shelley J, Cholleti A, Frye L, Greenwood J, Timlin M, Uchimaya M (2007) Epik: a software program for pKa prediction and protonation state generation for drug-like molecules. J Comput Aided Mol Des 21(12):681–691
Bayly CI, Cieplak P, Cornell W, Kollman PA (1993) A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J Phys Chem 97(40):10269–10280
Cornell WD, Cieplak P, Bayly CI, Kollmann PA (1993) Application of RESP charges to calculate conformational energies, hydrogen bond energies, and free energies of solvation. J Am Chem Soc 115(21):9620–9631
Dupradeau F-Y, Pigache A, Zaffran T, Savineau C, Lelong R, Grivel N, Lelong D, Rosanski W, Cieplak P (2010) The R.E.D. tools: advances in RESP and ESP charge derivation and force field library building. Phys Chem Chem Phys 12(28):7821–7839
Vanquelef E, Simon S, Marquant G, Garcia E, Klimerak G, Delepine JC, Cieplak P, Dupradeau F-Y (2011) R.E.D. Server: a web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucleic Acids Res 39(suppl 2):W511–W517
Dupradeau F-Y, Cézard C, Lelong R, Stanislawiak É, Pêcher J, Delepine JC, Cieplak P (2008) R.E.DD.B.: a database for RESP and ESP atomic charges, and force field libraries. Nucleic Acids Res 36(suppl 1):D360–D367
Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, Mackerell AD (2010) CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem 31(4):671–690
Li H, Raman CS, Glaser CB, Blasko E, Young TA, Parkinson JF, Whitlow M, Poulos TL (1999) Crystal structures of zinc-free and -bound heme domain of human inducible nitric-oxide synthase. J Biol Chem 274(30):21276–21284
Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26(16):1781–1802
MacKerell AD, Bashford D, Bellott, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 102(18):3586–3616
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14(1):33–38
Cho K-B, Derat E, Shaik S (2007) Compound I of nitric oxide synthase: the active site protonation state. J Am Chem Soc 129(11):3182–3188
Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA (2004) PDB2PQR: an automated pipeline for the setup of Poisson–Boltzmann electrostatics calculations. Nucleic Acids Res 32(suppl 2):W665–W667
Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA (2007) PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res 35(suppl 2):W522–W525
Li H, Robertson AD, Jensen JH (2005) Very fast empirical prediction and rationalization of protein pKa values. Proteins Struct Funct Bioinforma 61(4):704–721
Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) A smooth particle mesh Ewald method. J Chem Phys 103(19):8577–8593
Hoover WG (1985) Canonical dynamics: equilibrium phase-space distributions. Phys Rev A 31(3):1695
Feller SE, Zhang Y, Pastor RW, Brooks BR (1995) Constant pressure molecular dynamics simulation: the Langevin piston method. J Chem Phys 103(11):4613–4621
Nosé S (2002) A molecular dynamics method for simulations in the canonical ensemble. Mol Phys Int J Interface Chem Phys 100(1):191–198
Grest GS, Kremer K (1986) Molecular dynamics simulation for polymers in the presence of a heat bath. Phys Rev A 33(5):3628
DeLano WL (2002) The PyMOL molecular graphics system. DeLano Scientific, San Carlos
Volarea. João Ribeiro, Group of Computational BioChemistry, Faculty of Sciences of the University of Porto, Porto, Portugal, http://www.compbiochem.org/Software/Volarea/Home.html
Coleman RG, Sharp KA (2006) Travel depth, a new shape descriptor for macromolecules: application to ligand binding. J Mol Biol 362(3):441–458
Dijkstra EW (1959) A note on two problems in connexion with graphs. Numer Math 1(1):269–271
Delker SL, Ji H, Li H, Jamal J, Fang J, Xue F, Silverman RB, Poulos TL (2010) Unexpected binding modes of nitric oxide synthase inhibitors effective in the prevention of a cerebral palsy phenotype in an animal model. J Am Chem Soc 132(15):5437–5442
Aparna V, Desiraju GR, Gopalakrishnan B (2007) Insights into ligand selectivity in nitric oxide synthase isoforms: a molecular dynamics study. J Mol Graph Model 26(2):457–470
Rosenfeld RJ, Garcin ED, Panda K, Andersson G, Åberg A, Wallace AV, Morris GM, Olson AJ, Stuehr DJ, Tainer JA, Getzoff ED (2002) Conformational changes in nitric oxide synthases induced by chlorzoxazone and nitroindazoles: crystallographic and computational analyses of inhibitor potency. Biochemistry 41(47):13915–13925
Xue F, Li H, Delker SL, Fang J, Martasek P, Roman LJ, Poulos TL, Silverman RB (2010) Potent, highly selective, and orally bioavailable Gem-difluorinated monocationic inhibitors of neuronal nitric oxide synthase. J Am Chem Soc 132(40):14229–14238
Ji H, Delker SL, Li H, Martasek P, Roman LJ, Poulos TL, Silverman RB (2010) Exploration of the active site of neuronal nitric oxide synthase by the design and synthesis of pyrrolidinomethyl 2-aminopyridine derivatives. J Med Chem 53(21):7804–7824
Acknowledgments
This work has been partially supported by the Fundação para a Ciência e Tecnologia (FCT), Portugal (PTDC/QUI-QUI/121752/2010). We would like to thank F.-Y. Dupradeau for advice on using the R.E.D. server and for helpful discussions.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
The following supplementary material is available: Fig. S1: Sequence alignment of NOS oxygenase domains. Fig. S2 Chemical structures of 1 – 3 and torsions free to rotate during the docking process. Fig. S3 Description of the R.E.D. server procedure used to derive RESP charges for 1. Fig. S4 Close-up view of the lowest energy binding conformation of 3 inside eNOS. Fig. S5 Snapshot after 0 ns, 1.5 ns, 5 ns and 8 ns of MD simulation of the unbound iNOS isoform. Fig. S6 Average structures of 3:eNOSdimer and 3:eNOSmonomer calculated using the last 4 ns of simulation (A) and superimposition of these structures (B). Fig. S7 Overall RMSD of the protein backbone over 8 ns simulation is shown for the complexes 1:iNOS, 1:nNOS and 1:eNOS (A) and for inhibitor 1 (B). Fig. S8 Structural micro-environment around the active site of the unbound isoforms. Fig. S9 Superimposition of the X-ray structures of the 1:nNOS (violet, PDB ID 1K2R) and 1:eNOS (cyan, PDB ID 8NSE) complexes. Table S1 Summary of distances involved in the main interactions between inhibitor 1 and the neighboring amino acids residues on NOS isoforms. Fig. S10: Ability of Gln to switch between “open” and “closed” conformations creating an iNOS-specific subpocket. Fig. S11 Overlay of the average structure of 3 from the simulation of 3:nNOS and 3:eNOS and the conformations observed in the corresponding X-ray structures. (DOC 3868 kb)
Rights and permissions
About this article
Cite this article
Oliveira, B.L., Moreira, I.S., Fernandes, P.A. et al. Insights into the structural determinants for selective inhibition of nitric oxide synthase isoforms. J Mol Model 19, 1537–1551 (2013). https://doi.org/10.1007/s00894-012-1677-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-012-1677-8