Abstract
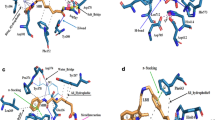
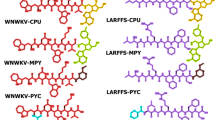
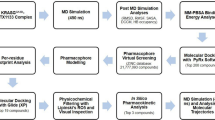
Among the many abnormally expressed proteins in ovarian cancer, the prominent cancer in women, ID1 (inhibitors of DNA binding protein 1) is a potential one among other several targets. Interaction of ID1 with ETS-1 (transcriptional activator of p16INK4a) suppresses the transcription of p16INK4a and causes abnormal cell proliferation. A peptide aptamer (ID1/3-PA7) has been designed to prevent this interaction and thereby leading to the transcription of p16INK4a. However, the structural basis behind the molecular interaction of ID1 with ETS-1 (agonist) and ID1/3-PA7 (antagonist) is poorly understood. In order to understand this structural recognition and their interaction mechanism, in silico methods were used. From this interaction analysis, the residues of ETS-1 involved in interaction with the p16INK4a promoter were found to be targeted by ID1. Subsequently, ETS-1 binding residues of ID1 were found to be targeted by its aptamer- ID1/3-PA7. These results suggest that both ETS-1 and ID1/3-PA7 binds at the same region harbored by the residues-H97, D100, R103, D104, L107, A144, C145, D149, D150 and C154 of ID1. All these observations correlate with the experimental reports, suggesting that the identified residues might play a crucial role in promulgating the oncogenic effects of ID1. In silico alanine scanning mutagenesis also confirms the role of identified hot spot residues in p16INK4a regulation. Finally, the molecular dynamic simulation studies reveal the prolonged stability of the aforementioned interacting complexes. The obtained results throw light on the structure and residues of ID1 involved in transcriptional regulation of p16INK4a.












Similar content being viewed by others
References
Parkin DM, Bray F, Ferlay J, Pisani P (2001) Estimating the world cancer burden: Globocan 2000. Int J Cancer 94(2):153–156. doi:10.1002/ijc.1440
Ohtani N, Zebedee Z, Huot TJ, Stinson JA, Sugimoto M, Ohashi Y, Sharrocks AD, Peters G, Hara E (2001) Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature 409(6823):1067–1070. doi:10.1038/35059131
Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H (1990) The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell 61(1):49–59
Biggs J, Murphy EV, Israel MA (1992) A human Id-like helix-loop-helix protein expressed during early development. Proc Natl Acad Sci U S A 89(4):1512–1516
Christy BA, Sanders LK, Lau LF, Copeland NG, Jenkins NA, Nathans D (1991) An Id-related helix-loop-helix protein encoded by a growth factor-inducible gene. Proc Natl Acad Sci U S A 88(5):1815–1819
Riechmann V, van Cruchten I, Sablitzky F (1994) The expression pattern of Id4, a novel dominant negative helix-loop-helix protein, is distinct from Id1, Id2 and Id3. Nucleic Acids Res 22(5):749–755. doi:10.1093/nar/22.5.749
Sun XH, Copeland NG, Jenkins NA, Baltimore D (1991) Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol 11(11):5603–5611
Davis RL, Cheng PF, Lassar AB, Weintraub H (1990) The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell 60(5):733–746
Ellenberger T, Fass D, Arnaud M, Harrison SC (1994) Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev 8(8):970–980. doi:10.1101/gad.8.8.970
Ma PC, Rould MA, Weintraub H, Pabo CO (1994) Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell 77(3):451–459
Murre C, McCaw PS, Baltimore D (1989) A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56(5):777–783
Henthorn P, Kiledjian M, Kadesch T (1990) Two distinct transcription factors that bind the immunoglobulin enhancer microE5/kappa 2 motif. Science 247(4941):467–470. doi:10.1126/science.2105528
Hu JS, Olson EN, Kingston RE (1992) HEB, a helix-loop-helix protein related to E2A and ITF2 that can modulate the DNA-binding ability of myogenic regulatory factors. Mol Cell Biol 12(3):1031–1042
Massari ME, Murre C (2000) Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol 20(2):429–440
Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, Cabrera CV, Buskin JN, Hauschka SD, Lassar AB et al. (1989) Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell 58(3):537–544
Skerjanc IS, Truong J, Filion P, McBurney MW (1996) A splice variant of the ITF-2 transcript encodes a transcription factor that inhibits MyoD activity. J Biol Chem 271(7):3555–3561. doi:10.1074/jbc.271.7.3555
Zhang Y, Babin J, Feldhaus AL, Singh H, Sharp PA, Bina M (1991) HTF4: a new human helix-loop-helix protein. Nucleic Acids Res 19(16):4555. doi:10.1093/nar/19.16.4555
Lassar AB, Davis RL, Wright WE, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H (1991) Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell 66(2):305–315. doi:10.1016/0092-8674(91)90620-E
srael MA, Hernandez MC, Florio M, Andres-Barquin PJ, Mantani A, Carter JH, Julin CM (1999) Id gene expression as a key mediator of tumor cell biology. Cancer Res 59(7 Suppl):1726s–1730s
Norton JD, Atherton GT (1998) Coupling of cell growth control and apoptosis functions of Id proteins. Mol Cell Biol 18(4):2371–2381
Norton JD (2000) ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J Cell Sci 113(Pt 22):3897–3905
Wibley J, Deed R, Jasiok M, Douglas K, Norton J (1996) A homology model of the Id-3 helix-loop-helix domain as a basis for structure-function predictions. Biochim Biophys Acta 1294(2):138–146
Kiewitz SD, Cabrele C (2005) Synthesis and conformational properties of protein fragments based on the Id family of DNA-binding and cell-differentiation inhibitors. Biopolymers 80(6):762–774. doi:10.1002/bip.20287
Garrell J, Modolell J (1990) The Drosophila extramacrochaetae locus, an antagonist of proneural genes that, like these genes, encodes a helix-loop-helix protein. Cell 61(1):39–48
Li R, Pei H, Watson DK (2000) Regulation of Ets function by protein - protein interactions. Oncogene 19(55):6514–6523. doi:10.1038/sj.onc.1204035
Perk J, Iavarone A, Benezra R (2005) Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer 5(8):603–614. doi:10.1038/nrc1673
Mern DS, Hasskarl J, Burwinkel B (2010) Inhibition of Id proteins by a peptide aptamer induces cell-cycle arrest and apoptosis in ovarian cancer cells. Br J Cancer 103(8):1237–1244. doi:10.1038/sj.bjc.6605897
Mern DS, Hoppe-Seyler K, Hoppe-Seyler F, Hasskarl J, Burwinkel B (2010) Targeting Id1 and Id3 by a specific peptide aptamer induces E-box promoter activity, cell cycle arrest, and apoptosis in breast cancer cells. Breast Cancer Res Treat 124(3):623–633. doi:10.1007/s10549-010-0810-6
Lin CQ, Singh J, Murata K, Itahana Y, Parrinello S, Liang SH, Gillett CE, Campisi J, Desprez PY (2000) A role for Id-1 in the aggressive phenotype and steroid hormone response of human breast cancer cells. Cancer Res 60(5):1332–1340
Schindl M, Oberhuber G, Obermair A, Schoppmann SF, Karner B, Birner P (2001) Overexpression of Id-1 protein is a marker for unfavorable prognosis in early-stage cervical cancer. Cancer Res 61(15):5703–5706
Langlands K, Down GA, Kealey T (2000) Id proteins are dynamically expressed in normal epidermis and dysregulated in squamous cell carcinoma. Cancer Res 60(21):5929–5933
Maruyama H, Kleeff J, Wildi S, Friess H, Buchler MW, Israel MA, Korc M (1999) Id-1 and Id-2 are overexpressed in pancreatic cancer and in dysplastic lesions in chronic pancreatitis. Am J Pathol 155(3):815–822. doi:10.1016/S0002-9440(10)65180-2
Ouyang XS, Wang X, Lee DT, Tsao SW, Wong YC (2002) Over expression of ID-1 in prostate cancer. J Urol 167(6):2598–2602. doi:10.1016/S0022-5347(05)65044-6
Zhang X, Ling MT, Feng H, Wong YC, Tsao SW, Wang X (2004) Id-I stimulates cell proliferation through activation of EGFR in ovarian cancer cells. Br J Cancer 91(12):2042–2047. doi:10.1038/sj.bjc.6602254
Roy A, Kucukural A, Zhang Y (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5(4):725–738. doi:10.1038/nprot.2010.5
Zhang Y (2007) Template-based modeling and free modeling by I-TASSER in CASP7. Proteins 69(Suppl 8):108–117. doi:10.1002/prot.21702
McGuffin LJ, Bryson K, Jones DT (2000) The PSIPRED protein structure prediction server. Bioinformatics 16(4):404–405. doi:10.1093/bioinformatics/16.4.404
Hess B, Kutzner C, van der Spoel D, Lindahl E (2008) GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J Chem Theor Comput 4(3):435–447. doi:10.1021/ct700301q
Van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ (2005) GROMACS: fast, flexible, and free. J Comput Chem 26(16):1701–1718. doi:10.1002/jcc.20291
Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE (2010) Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78(8):1950–1958. doi:10.1002/prot.22711
Kawata M, Nagashima U (2001) Particle mesh Ewald method for three-dimensional systems with two-dimensional periodicity. Chem Phys Lett 340(1–2):165–172. doi:10.1016/S0009-2614(01)00393-1
Hess B, Bekker H, Berendsen HJC, Fraaije JGEM (1997) LINCS: A linear constraint solver for molecular simulations. J Comput Chem 18(12):1463–1472. doi:10.1002/(sici)1096-987x(199709)18:12<1463::aid-jcc4>3.0.co;2-h
Miyamoto S, Kollman PA (1992) Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J Comput Chem 13(8):952–962. doi:10.1002/jcc.540130805
Martonak R, Laio A, Parrinello M (2003) Predicting crystal structures: the Parrinello-Rahman method revisited. Phys Rev Lett 90(7):075503. doi:10.1103/PhysRevLett.90.075503
Goyal A, Muthu K, Panneerselvam M, Pole AK, Ramadas K (2011) Molecular dynamics simulation of the Staphylococcus aureus YsxC protein: molecular insights into ribosome assembly and allosteric inhibition of the protein. J Mol Model 17(12):3129–3149. doi:10.1007/s00894-011-0998-3
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 26(2):283–291. doi:10.1107/S0021889892009944
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2(9):1511–1519. doi:10.1002/pro.5560020916
Dominguez C, Boelens R, Bonvin AM (2003) HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc 125(7):1731–1737. doi:10.1021/ja026939x
de Vries SJ, van Dijk AD, Krzeminski M, van Dijk M, Thureau A, Hsu V, Wassenaar T, Bonvin AM (2007) HADDOCK versus HADDOCK: new features and performance of HADDOCK2.0 on the CAPRI targets. Proteins 69(4):726–733. doi:10.1002/prot.21723
Lamber EP, Vanhille L, Textor LC, Kachalova GS, Sieweke MH, Wilmanns M (2008) Regulation of the transcription factor Ets-1 by DNA-mediated homo-dimerization. EMBO J 27(14):2006–2017. doi:10.1038/emboj.2008.117
Garvie CW, Pufall MA, Graves BJ, Wolberger C (2002) Structural analysis of the autoinhibition of Ets-1 and its role in protein partnerships. J Biol Chem 277(47):45529–45536. doi:10.1074/jbc.M206327200
Wallace AC, Laskowski RA, Thornton JM (1995) LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng 8(2):127–134. doi:10.1093/protein/8.2.127
Manivel P, Muthukumaran J, Kannan M, Krishna R (2011) Insight into residues involved in the structure and function of the breast cancer associated protein human gamma synuclein. J Mol Model 17(2):251–263. doi:10.1007/s00894-010-0718-4
Kruger DM, Gohlke H DrugScorePPI webserver: fast and accurate in silico alanine scanning for scoring protein-protein interactions. Nucleic Acids Res 38(Web Server issue):W480–W486. doi: 10.1093/nar/gkq471
Pesce S, Benezra R (1993) The loop region of the helix-loop-helix protein Id1 is critical for its dominant negative activity. Mol Cell Biol 13(12):7874–7880
Yokota Y, Mori S (2002) Role of Id family proteins in growth control. J Cell Physiol 190(1):21–28. doi:10.1002/jcp.10042
Bounpheng MA, Dimas JJ, Dodds SG, Christy BA (1999) Degradation of Id proteins by the ubiquitin-proteasome pathway. FASEB J 13(15):2257–2264
Deed RW, Armitage S, Norton JD (1996) Nuclear localization and regulation of Id protein through an E protein-mediated chaperone mechanism. J Biol Chem 271(39):23603–23606. doi:10.1074/jbc.271.39.23603
Weiss MS, Brandl M, Suhnel J, Pal D, Hilgenfeld R (2001) More hydrogen bonds for the (structural) biologist. Trends Biochem Sci 26(9):521–523
Lesk A (2001) Introduction to protein architecture. Oxford University Press, Oxford
Guo Z, Mohanty U, Noehre J, Sawyer TK, Sherman W, Krilov G (2010) Probing the alpha-helical structural stability of stapled p53 peptides: molecular dynamics simulations and analysis. Chem Biol Drug Des 75(4):348–359. doi:10.1111/j.1747-0285.2010.00951.x
Gallivan JP, Dougherty DA (1999) Cation-pi interactions in structural biology. Proc Natl Acad Sci U S A 96(17):9459–9464. doi:10.1073/pnas.96.17.9459
Acknowledgments
R. Krishna and P. Manivel thanks University Grant Commission (UGC), Government of India for providing financial assistance (F.No.37-313/2009 (SR)) to carry out the Research work. Kannan M. (No.F. 14-2(SC)/2009 (SA-III)) and Nishith Saurav Topno (No.F. 14-2(ST)/2010 (SA-III)) thank UGC for Rajiv Gandhi National Fellowship to pursue their PhD degree. J. Muthukumaran (No.09/559/(0076)/2011/EMR-I) thanks CSIR (Council of Scientific and Industrial Research) for Senior Research fellowship. R. Krishna also thanks Department of Biotechnology and Department of Information technology, Government of India, New Delhi for their financial support to Centre for Bioinformatics, Pondicherry University. We thank E. Malanco, P. Elavarasan and S. Manikandan, Senior technical assistants, Centre for Bioinformatics, Pondicherry University for their technical supports. Authors also extend their greetings and regards to Jayakanthan M. and Bhuvaenshwari S. for their timely help to carry out the research work.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 313 kb)
Rights and permissions
About this article
Cite this article
Muthu, K., Panneerselvam, M., Jayaraman, M. et al. Structural insights into interacting mechanism of ID1 protein with an antagonist ID1/3-PA7 and agonist ETS-1 in treatment of ovarian cancer: molecular docking and dynamics studies. J Mol Model 18, 4865–4884 (2012). https://doi.org/10.1007/s00894-012-1489-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-012-1489-x