Abstract
The design and development of antioxidant molecules have lately gained a great deal of focus which is attributed to their immense biomedicinal importance in combating the free radical associated health hazards. In a situation to replenish the endogenous antioxidant loss, synthetic molecules with potent antioxidant activity is demanded. The present work thus aims at in silico modeling of antioxidant molecules that may facilitate in searching and designing of new chemical entities with enhanced activity profile. A series of cinnamic acid and caffeic acid derivatives having the ability to inhibit lipid peroxidation have been modeled in the present work. Three different types of models were developed using different chemometric and cheminformatics tools to identify the essential structural attributes: (a) descriptor based QSAR models, (b) 3D pharmacophore models and (c) HQSAR (hologram QSAR) models. For the conventional QSAR modeling, descriptors belonging to different categories [quantum chemical descriptors (Mulliken charges of the common atoms of the molecules), thermodynamic descriptors, electronic descriptors, structural descriptors and spatial descriptors] were calculated for the development of statistically significant as well as well interpretable quantitative structure-activity relationship (QSAR) models. Two different chemometric tools [genetic function approximation (GFA) and genetic partial least squares (G/PLS)] were employed for the development of the QSAR models. The 3D pharmacophore model focused on the essential pharmacophoric features while the HQSAR model implicated the prime structural fragments that were necessitated for the optimal anti-lipid peroxidative activity of the molecules. All the models were validated based on internal, external and overall validation statistics. Randomization was performed in order to ensure the absence of chance correlation in the developed models. Among all models, the descriptor-based model developed using the GFA-spline technique yielded the most satisfactory results. The results obtained from all the models corroborate well with each other and chiefly signify the importance of the ketonic oxygen of the amide/ acid fragment and the ethereal oxygen substituted on the parent phenyl ring of the molecules under study. Thus the models can efficiently be utilized for extensive screening of large datasets and their subsequent activity prediction.

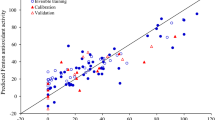
Schematic diagram showing different features at various positions favouring the anti-lipid peroxidative activity profile of the molecules obtained using different QSAR techniques







Similar content being viewed by others
References
Pacher P, Beckman JS, Liaudet L (2007) Nitric oxide and peroxynitrite in health and disease. Physiol Rev 87:315–424
Dizdaroglu M, Jaruga P, Birincioglu M, Rodriguez H (2002) Free radical-induced damage to DNA: mechanisms and measurement. Free Radic Biol Med 32:1102–1115
Dreher D, Junod AF (1996) Role of oxygen free radicals in cancer development. Eur J Cancer 32:30–38
Leonard SS, Wang S, Shi X, Jordan BS, Castranova V, Dubick MA (2000) Wood smoke particles generate free radicals and cause lipid peroxidation, DNA damage, NFkB activation and TNF-a release in macrophages. Toxicology 150:147–157
Halliwell B, Gutteridge JMC (1990) Role of free radicals and catalytic metal ions in human disease: an overview. Method Enzymol 186:1–85
Winrow VR, Winyard PG, Morris CJ, Blake DR (1993) Free radicals in inflammation: second messengers and mediators of tissue destruction. Br Med Bull 49:506–522
Leopold JA, Loscalzo J (2009) Oxidative risk for atherothrombotic cardiovascular disease. Free Radic Biol Med 47:1673–1706
Frei B, Stocker R, Ames BN (1988) Antioxidant defenses and lipid peroxidation in human blood plasma. Proc Natl Inst Sci India 85:9748–9752
Gey KF, Brubacher GB, Stahelin HB (1987) Plasma levels of antioxidant vitamins in relation to ischemic heart disease and cancer. Am J Clin Nutr 45:1368–1377
Block G, Patterson B, Subar A (1992) Fruit, vegetables and cancer prevention: a review of the epidemiological evidence. Nutr Cancer 18:1–29
Kritchevisky SB (1999) Beta-carotene, carotenoids and the prevention of coronary heart disease. J Nutr 129:5–8
Buchwald P, Bodor N (2002) Computer-aided drug design: the role of quantitative structure–property, structure–activity and structure–metabolism relationships (QSPR, QSAR, QSMR). Drugs Future 27:577–588
Todeschini R, Consonni V (2000) Handbook of molecular descriptors. Wiley-VCH, Weinheim
Worachartcheewan A, Nantasenamat C, Isarankura-Na-Ayudhya C, Prachayasittikul S, Prachayasittikul V (2011) Predicting the free radical scavenging activity of curcumin derivatives. Chemom Intell Lab Syst 109:207–216
Li YW, Li B, He J, Qian P (2011) Quantitative structure–activity relationship study of antioxidative peptide by using different sets of amino acids descriptors. J Mol Struct 998:53–61
Nikitakis A, Kourounakis AP (2011) QSAR of substituted morpholines with antioxidant and squalene synthase inhibitory activity. Med Chem Res 20:566–575
Mitra I, Saha A, Roy K (2011) Development of multiple QSAR models for consensus predictions and unified mechanistic interpretations of the free-radical scavenging activities of chromone derivatives. J Mol Model http://dx.doi.org/10.1007/s00894-011-1198-x
Mitra I, Saha A, Roy K (2010) Pharmacophore mapping of arylamino-substituted benzo[b]thiophenes as free radical scavengers. J Mol Model 16:1585–1596
Jung YS, Kang TS, Yoon JH, Joe BY, Lim HJ, Seong CM, Park WK, Kong JY, Chob J, Park NS (2002) Synthesis and evaluation of 4-hydroxyphenylacetic acid amides and 4-hydroxycinnamamides as antioxidants. Bioorg Med Chem Lett 12:2599–2602
Kang TS, Jo HO, Park WK, Kim JP, Konishi Y, Kong JY, Park NS, Jung YS (2008) Synthesis and antioxidant activities of 3,5-dialkoxy-4-hydroxycinnamamides. Bioorg Med Chem Lett 18:1663–1667
Rajan P, Vedernikova I, Cos P, Berghe DV, Augustyns K, Haemers A (2001) Synthesis and evaluation of caffeic acid amides as antioxidants. Bioorg Med Chem Lett 11:215–217
Semichem Inc (2003) GaussView3.0, Gaussian Inc, Pittsburgh, PA
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Vreven JT, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA (2003) Gaussian 03, Revision B.05, Gaussian Inc, Pittsburgh, PA
Accelrys Inc (2005) Cerius2 Version 4.10. Accelrys Inc. San Diego, CA
Dougherty ER, Barrera J, Brun M, Kim S, Cesar RM, Chen Y, Bittner M, Trent JM (2002) Inference from clustering with application to gene-expression microarrays. J Comput Biol 9:105–126
Everitt B, Landau S, Leese M (2001) Cluster analysis. Arnold, London
SPSS Inc. (2011) SPSS. SPSS Inc, Chicago. http://www.spss.com
Rogers D, Hopfinger AJ (1994) Application of genetic function approximation to quantitative structure–activity relationships and quantitative structure–property relationships. J Chem Inf Comput Sci 34:854–866
Wold S, Sjostrom M, Eriksson L (2001) PLS-regression: a basic tool of chemometrics. Chemom Intell Lab Syst 58:109–130
Smellie A, Teig SL, Towbin P (1995) Poling: promoting conformational variation. J Comput Chem 16:171–187
Accelrys Inc (2010) Discovery Studio 2.1. Accelrys Inc, San Diego, CA
Sutter J, Guner OF, Hoffman R, Li H, Waldman M (2000) HypoGen: an automated system for generating 3D predictive pharmacophore models. In: Guner OF (ed) Pharmacophore perception, development, and use in drug design. International University Line, La Jolla, pp 501–511
Tong W, Lowis DR, Perkins R, Chen Y, Welsh WJ, Goddette DW, Heritage TW, Sheehan DM (1998) Evaluation of quantitative structure–activity relationship methods for large-scale prediction of chemicals binding to the estrogen receptor. J Chem Inf Comput Sci 38:669–677
Waller CL (2004) A comparative QSAR study using CoMFA, HQSAR, and FRED/SKEYS paradigms for estrogen receptor binding affinities of structurally diverse compounds. J Chem Inf Comput Sci 44:758–765
SYBYL 7.3 (2006) Tripos Inc, St. Louis. www.tripos.com
Wold S, Johansson E, Cocchi M (1993) PLS: partial least squares projections to latent structures. In: Kubiniyi H (ed) 3D QSAR in drug design: theory methods and applications. ESCOM, Leiden, pp 523–550
Ojha PK, Mitra I, Das RN, Roy K (2011) Further exploring r 2m metrics for validation of QSPR models. Chemom Intell Lab Syst 107:194–205
Roy K, Mitra I, Kar S, Ojha P, Das RN, Kabir H (2012) Comparative studies on some metrics for external validation of QSPR models. J Chem Inf Model 52:396–408
Golbraikh A, Tropsha A (2002) Beware of q2! J Mol Graph Model 20:269–276
Mitra I, Saha A, Roy K (2010) Exploring quantitative structure–activity relationship (QSAR) studies of antioxidant phenolic compounds obtained from traditional Chinese medicinal plants. Mol Simul 36:1067–1079
Toropova AP, Toropov AA, Martyanov SE, Benfenati E, Gini G, Leszczynska D, Leszczynski J (2012) CORAL: QSAR modeling of toxicity of organic chemicals towards Daphnia magna. Chemom Intell Lab Syst 110:177–181
Dashtbozorgi Z, Golmohammadi H (2010) Prediction of air to liver partition coefficient for volatile organic compounds using QSAR approaches. Eur J Med Chem 45:2182–2190
CORAL (2011) CORAL freeware available at http://www.insilico.eu/coral
Wright JS, Johnson ER, Dilabio GA (2001) Predicting the activity of phenolic antioxidants: theoretical method, analysis of substituent effects, and application to major families of antioxidants. J Am Chem Soc 123:1173–1183
Patrick GL (2009) An introduction to medicinal chemistry. Oxford University Press, New York
Pietta PG (2000) Flavonoids as antioxidants. J Nat Prod 63:1035–1042
Acknowledgments
This research work is supported in the form of a major research project to KR by Indian Council of Medical Research (ICMR), New Delhi and a senior research fellowship to IM by Council of Scientific & Industrial Research (CSIR), New Delhi.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mitra, I., Saha, A. & Roy, K. In silico development, validation and comparison of predictive QSAR models for lipid peroxidation inhibitory activity of cinnamic acid and caffeic acid derivatives using multiple chemometric and cheminformatics tools. J Mol Model 18, 3951–3967 (2012). https://doi.org/10.1007/s00894-012-1392-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-012-1392-5