Abstract
Isoniazid (INH) is a front-line drug used in the treatment of tuberculosis (TB), a disease that remains a major cause of death worldwide. Isoniazid is a prodrug, requiring activation in the mycobacterial cell by the catalase-peroxidase (CP) enzyme. Recent studies have suggested that acetylation of INH by the arylamine-N-acetyltransferase from Mycobacterium tuberculosis (TBNAT) may be a possible cause of inactivation of the drug thus resulting in resistant strains. In this study, computational techniques were applied to investigate the binding of isoniazid to three TBNAT isoforms: wild type, G68R and L125M. Since there is no experimental structure available, molecular dynamics (MD) simulations were initially used for the refinement of TBNAT homology models. Distinct conformations of the models were selected during the production stage of MD simulations for molecular docking experiments with the drug. Finally, each mode of binding was refined by new molecular MD simulations. Essential dynamics (ED) analysis and linear interaction energy calculations (LIE) were used to evaluate the impact of amino acid substitutions on the structural and binding properties of the enzymes. The results suggest that the wild type and the G68R TBNATs have a similar pattern of affinity to INH. On the other hand, the calculated enzyme-INH dissociation constant (KD) was estimated 33 times lower for L125M isoform in comparison with wild type enzyme. This last finding is consistent with the hypothesis that isolated mutations in the tbnat gene can produce M. tuberculosis strains resistant to isoniazid.

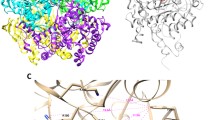
Global structure of the complexes and binding mode of INH calculated for the last frame of MD simulations






Similar content being viewed by others
Abbreviations
- TB:
-
Tuberculosis
- INH:
-
Isoniazid
- CP:
-
Catalase-peroxidase
- NAT:
-
Arylamine N-acetyltransferase
- TBNAT:
-
Arylamine-N-acetyltransferase from Mycobacterium tuberculosis
- MD:
-
Molecular dynamics
- ED:
-
Essential dynamics
- LIE:
-
Linear interaction energy
- SNPs:
-
Single nucleotide polymorphisms
- K D :
-
Dissociation constant
- NCBI:
-
National Center for Biotechnology Information
- InChITM :
-
The IUPAC International Chemical Identifier
- AM1:
-
Austin model 1
- MEP:
-
Molecular electrostatic potential
- CHELPG:
-
Charges from electrostatic grid based
- SPC:
-
Simple point charge
- ADT:
-
AutoDock tools
- RMSD:
-
Root mean square deviation
- LGA:
-
Lamarckian genetic algorithm
- LS:
-
Local search
- vdW:
-
van der Waals
- el:
-
Electrostatic
- Rg:
-
Radius of gyration
- NHb:
-
Number of intramolecular hydrogen bonds
- SASA:
-
Surface accessible solvent area
- RMSF:
-
Root mean square fluctuation
- MC:
-
Monte Carlo
References
Zhang Y, Young DB (1993) Molecular mechanisms of isoniazid: a drug at the front line of tuberculosis control. Trends Microbiol 1:109–113
Timmins GS, Deretic V (2006) Mechanisms of action of isoniazid. Mol Microbiol 62:1220–1227
Balcells ME, Thomas SL, Godfrey-Faussett P, Grant AD (2006) Isoniazid preventive therapy and risk for resistant tuberculosis. Emerg Infect Dis 12:744–751
WHO (2011) World Health Organization. http://www.who.int/infectious-disease-report/pages/ch2text.html. Accessed 02 April 2011
Bhakta S, Besra GS, Upton AM, Parish T, Vernon CS, Gibson KJC, Knutton S, Gordon S, Silva RP, Anderton MC, Sim E (2004) Arylamine N-acetyltransferase is required for synthesis of mycolic acids and complex lipids in Mycobacterium bovis BCG and represents a novel drug target. J Exp Med 199:1191–1199
Sandy J, Mushtaq A, Holton SJ, Schartau P, Noble MEM, Sim E (2005) Investigation of the catalytic triad of arylamine N-acetyltransferases: essential residues required for acetyl transfer to arylamines. Biochem J 390:115–123
Mdluli K, Swanson J, Fischer E, Lee RE, Barry CE (1998) Mechanisms involved in the intrinsic isoniazid resistance of Mycobacterium avium. Mol Microbiol 27:1223–1233
Somoskovi A, Parsons LM, Salfinger M (2001) The molecular basis of resistance to isoniazid, rifampin, and pyrazinamide in Mycobacterium tuberculosis. Respir Res 2:164–168
Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um KS, Wilson T, Collins D, Lisle G, Jacobs WR Jr (1994) inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263:227–230
Kelley CL, Rouse DA, Morris SL (1997) Analysis do ahpC gene mutations in isoniazid resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Ch 41:2057–2058
Ramaswamy S, Musser JM (1998) Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber Lung Dis 79:3–29
Mdluli K, Slayden RA, Zhu Y, Ramaswamy S, Pan X, Mead D, Crane DD, Musser JM, Barry CE (1998) Inhibition of a Mycobacterium tuberculosis beta-ketoacyl ACP synthase by isoniazid. Science 280:1607–1610
Lee ASG, Teo ASM, Wong SY (2001) Novel mutations in ndh in isoniazid-resistant Mycobacterium tuberculosis isolates. Antimicrob Agents Ch 45:2157–2159
Almeida Da Silva PE, Palomino JC (2011) Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: classical and new drugs. J Antimicrob Chemother 66:1417–1430
van Rie A, Warren R, Mshanga I, Jordaan AM, van der Spuy GD, Richardson M, Simpson J, Gie RP, Enarson DA, Beyers N, van Helden PD, Victor TC (2001) Analysis for a limited number of gene codons can predict drug resistance of Mycobacterium tuberculosis in a high-incidence community. J Clin Microbiol 39:636–641
Silva MSN, Senna S, Ribeiro MO, Valim AM, Telles MA, Kritski AL, Morlock GP, Cooksey RC, Zaha A, Rossetti MLR (2003) Mutations in katG, inhA and ahpC genes of brazilian isoniazid-resistant isolates of Mycobacterium tuberculosis. J Clin Microbiol 41:4471–4474
Dalla Costa ER, Ribeiro MO, Silva MSN, Arnold LS, Rostirolla DC, Cafrune PI, Espinoza RC, Palaci M, Telles M, Ritacco V, Suffys PN, Lopes ML, Campelo CL, Miranda SS, Kremer K, Silva Almeida PE, Fonseca LS HOJL, Kritski AL, Rossetti MLR (2009) Correlations of mutations in katG, oxyR-ahpC and inhA genes and in vitro susceptibility in Mycobacterium tuberculosis clinical strains segregated by spoligotype families from tuberculosis prevalent countries in South America. BMC Microbiol 9:1–11
Coelho MB, Dalla Costa ER, Vasconcellos SEG, Linck N, Ramos RM, de Amorim HLN, Suffys PN, Santos AR, da Silva PEA, Ramos DF, Silva MSN, Rossetti MLR (2011) Sequence and structural characterization of tbnat gene in isoniazid-resistant Mycobacterium tuberculosis: identification of new mutations. Mutat Res 712:33–39
Sim E, Walters K, Boukouvala S (2008) Arylamine N-acetyltransferases: from structure to function. Drug Metab Rev 40:479–510
Weber WW, Hein DW (1985) N-acetylation pharmacogenetics. Pharmacol Rev 37:25–27
Sim E, Lack N, Wang CJ, Long H, Westwood I, Fullam E, Kawamura A (2008) Arylamine N-acetyltransferases: structural and functional implications of polymorphisms. Toxicology 254:170–183
Upton AM, Mushtaq A, Victor TC, Sampson SL, Sandy J, Smith DM, van Helden PD, Sim E (2001) Arylamine N-acetyltransferase of Mycobacterium tuberculosis is a polymorphic enzyme and a site of isoniazid metabolism. Mol Microbiol 42:309–317
Sandy J, Holton S, Fullam E, Sim E, Noble M (2005) Binding of the anti-tubercular drug isoniazid to the arylamine N-acetyltransferase protein from Mycobacterium smegmatis. Protein Sci 14:775–782
Payton M, Auty R, Delgoda R, Everett M, Sim E (1999) Cloning and characterization of arylamine N-acetyltransferase genes from Mycobacterium smegmatis and Mycobacterium tuberculosis: increased expression results in isoniazid resistance. J Bacteriol 181:1343–1347
Sholto-Douglas-Vernon C, Sandy J, Victor TC, Sim E, Helden PD (2005) Mutational and expression analysis of tbnat and its response to isoniazid. J Med Microbiol 54:1189–1197
Coelho MB (2008) Análise da presença de bombas de efluxo e mutações no gene nat em Mycobacterium tuberculosis resistentes a isoniazida. Universidade Luterana do Brasil, Dissertação de Mestrado
Fullam E, Westwood IM, Anderton MC, Lowe ED, Sim E, Noble ME (2008) Divergence of cofator recognition across evolution: coenzyme A binding in a prokaryotic arylamine N-acetyltransferase. J Mol Biol 375:178–191
Dalby A, Nourse JG, Hounshell WD, Gushurst AKI, Grier DL, Leland BA, Laufer J (1992) Description of several chemical structure file formats used by computer programs developed at Molecular Design Limited. J Chem Inf Comp Sci 32:244–255
Spessard GO (1998) ACD Labs/LogP dB 3.5 and ChemSketch 3.5. J Chem Inf Comput Sci 38:1250–1253
Thompson MA (2004) Planaria Software LLC, Seattle, WA, http://www.arguslab.com
Dewar MJS, Zoebisch EG, Healy EF, Stewart JJP (1985) AM1: a new general purpose quantum mechanical molecular model. J Am Chem Soc 107:3902–3909
Aalten DMF, Bywater R, Findlay JBC, Hendlich M, Hooft RWW, Vriend G (1996) PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. J Comp Aid Molec Des 10:255–262
Chuang IL, Gershenfeld N, Kubinec M (1998) Experimental implementation of fast quantum searching. Phys Rev Lett 80:3408–3411
Breneman CM, Wiberg KB (1990) Determining atom-centered monopoles from molecular electrostatic potentials. The need for high sampling density in formamide conformational analysis. J Comput Chem 11:361–373
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA (2004) Gaussian 03 Revision C.02. Gaussian Inc, Wallingford, CT http://www.gaussian.com/home.htm
Oostenbrink C, Villa A, Mark AE, van Gunsteren WF (2004) A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J Comput Chem 25:1656–1676
van der Spoel D, Lindahl E, Hess B, van Buuren AR, Apol E, Meulenhoff PJ, Tieleman DP, Sijbers ALTM, Feenstra KA, van Drunen R, Berendsen HJC (2010) Gromacs User Manual version 4.5.4 http://www.gromacs.org
Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J (1981) Intermolecular forces. In: Pullman B (ed) Interaction models for water in relation to protein hydration. Reidel, Dordrecht, pp 331–342
Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690
Nosé S, Klein ML (1983) Constant pressure molecular dynamics for molecular systems. Mol Phys 50:1055–1076
Parrinello M, Rahman A (1981) Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys 52:7182–7190
Hess B, Bekker H, Berendsen HJC, Fraaije JGEM (1997) LINCS: A linear constraint solver for molecular simulations. J Comp Chem 18:1463–1472
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an Nlog-(N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092
Goodsell DS, Morris GM, Olson AJ (1996) Automated docking of flexible ligands: applications of AutoDock. J Mol Recognit 9:1–5
Goodsell DS (2005) Computational docking of biomolecular complexes with AutoDock. In: Golemis E, Adams P (eds) Protein-protein interactions: a molecular cloning manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, p 885
Morris GM, Huey R, Olson AJ (2008) Using AutoDock for ligand-receptor docking. Curr Prot Bioinf 24:8.14.1–8.14.40
Sanner MF (1999) Python: a programming language for software integration and development. J Mol Graphics Mod 17:57–61
Gasteiger J, Marsili M (1980) Iterative partial equalization of orbital electronegativity - a rapid access to atomic charges. Tetrahedron 36:3219–3228
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ (1998) Automated docking using a lamarckian genetic algorithm and and empirical binding free energy function. J Comput Chem 19:1639–1662
Solis FJ, Wets RJB (1981) Minimization by random search techniques. Math Oper Res 6:19–30
Åqvist J, Medina C, Samuelsson JE (1994) A new method for predicting binding affinity in computer-aided drug design. Protein Eng 7:385–391
Hansson T, Marelius J, Aqvist J (1998) Ligand binding affinity prediction by linear interaction energy methods. J Comput Aided Mol Des 12:27–35
Wallace AC, Laskowski RA, Thornton JM (1996) LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng 8:127–134
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723
Payton M, Gifford C, Schartau P, Hagemeier C, Mushtaq A, Lucas S, Pinter K, Sim E (2001) Evidence towards the role of arylamine N-acetyltransferase in Mycobacterium smegmatis and development of a specific antiserum against the homologous enzyme of Mycobacterium tuberculosis. Microbiology 147:3295–3302
Payton MA, Sim E (1998) Genotyping human arylamine N-acetyltransferase type 1 (NAT1): the identification of two novel allelic variants. Biochem Pharmacol 55:361–366
Mushtaq A, Payton M, Sim E (2002) The COOH terminus of arylamine N-acetyltransferase from Salmonella typhimurium controls enzymic activity. J Biol Chem 277:12175–12181
Delgoda R, Lian LY, Sandy J, Sim E (2003) NMR investigation of the catalytic mechanism of arylamine N-acetyltransferase from Salmonella typhimurium. Biochim Biophys Acta 1620:8–14
Sikora AL, Frankel BA, Blanchard JS (2008) Kinetic and chemical mechanism of arylamine N-acetyltransferase from Mycobacterium tuberculosis. Biochemistry 47:10781–9
Aqvist J, Medina C, Samuelsson JE (1994) A new method for predicting binding affinity in computer-aided drug design. Protein Eng 7:385–391
de Amorim HL, Caceres RA, Netz PA (2008) Linear interaction energy (LIE) method in lead discovery and optimization. Curr Drug Targets 9:1100–1105
Wang JP, Morin P, Wang W, Kollman PAJ (2001) Use of MM-PBSA in reproducing the binding free energies to HIV-1 RT of TIBO derivatives and predicting the binding mode to HIV-1 RT of efavirenz by docking and MM-PBSA. Am Chem Soc 123:5221–5230
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ramos, R.M., Perez, J.M., Baptista, L.A. et al. Interaction of wild type, G68R and L125M isoforms of the arylamine-N-acetyltransferase from Mycobacterium tuberculosis with isoniazid: a computational study on a new possible mechanism of resistance. J Mol Model 18, 4013–4024 (2012). https://doi.org/10.1007/s00894-012-1383-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-012-1383-6