Abstract
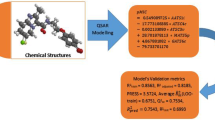
The virtual combinatorial chemistry approach as a methodology for generating chemical libraries of structurally-similar analogs in a virtual environment was employed for building a general mixed virtual combinatorial library with a total of 53.871 6-FQ structural analogs, introducing the real synthetic pathways of three well known 6-FQ inhibitors. The druggability properties of the generated combinatorial 6-FQs were assessed using an in-house developed drug-likeness filter integrating the Lipinski/Veber rule-sets. The compounds recognized as drug-like were used as an external set for prediction of the biological activity values using a neural-networks (NN) model based on an experimentally-determined set of active 6-FQs. Furthermore, a subset of compounds was extracted from the pool of drug-like 6-FQs, with predicted biological activity, and subsequently used in virtual screening (VS) campaign combining pharmacophore modeling and molecular docking studies. This complex scheme, a powerful combination of chemometric and molecular modeling approaches provided novel QSAR guidelines that could aid in the further lead development of 6-FQs agents.






Similar content being viewed by others
Abbreviations
- TB:
-
Tuberculosis
- ATP:
-
Adenosine triphosphate
- MIC:
-
Minimal Inhibitory Concentration
- 6-FQs:
-
6-Fluoroquinolones
- SAR:
-
Structure-Activity Relationships
- QSAR:
-
Quantitative Structure-Activity Relationships
- CombiChem:
-
Combinatorial Chemistry
- SSS:
-
Substructure Search
- NN:
-
Neural-Networks
- KANN:
-
Kohonen Artificial Neural Networks
- CP ANN:
-
Counter-Propagation Artificial Neural Networks
- GHA:
-
Global Hypothetical Activity
- LBP:
-
Ligand-Based Pharmacophore
- SBP:
-
Structure-Based Pharmacophore
- VS:
-
Virtual Screening
References
Munro SA, Lewin SA, Smith HJ, Engel ME, Feetheim A, Volmink J (2007) Patient adherence to tuberculosis treatment: a systematic review of qualitative research. PloS Med 4:1230–1245
Du Toit LC, Pillay V, Danckwerts MP (2006) Tuberculosis chemotherapy: current drug delivery spproaches. Respir Res 7:118–136
Bhanu NV, van Soolingen D, van Embden JDA, Seth P (2004) Two Mycobacterium fortuitum strains isolated from pulmonary tuberculosis patients in Delhi IS6110 harbour homologue. Diagn Micro Infec Dis 48:107–110
Dussurget O, Rodriguez M, Smith I (1998) Protective role of Mycobacterium smegmatis IdeR against reactive oxygen species and isoniazid toxicity. Tubercle Lung Dis 79:99–106
Field SK, Fisher D, Cowie RL (2004) Mycobacterium avium complex pulmonary disease in patients without HIV infection. Chest 126:566–581
Zhang Y, Martens KP, Denkin S (2006) New drug candidates and therapeutic targets for tuberculosis therapy. Drug Discovery Today 11:21–27
Wigley DB (1995) Structure and mechanism of DNA gyrase. In: Eckstein F, Lilley DMJ (eds) Nucleic Acids Molecular Biol. Springer, Berlin, pp 165–176
Barnard FM, Maxwell A (2001) Interaction between DNA gyrase and quinolones: effects of alanine mutations at GyrA subunit residues Ser83 and Asp87. Antimicrob Agents Chemother 45:1994–2000
Peng H, Marians KJ (1993) Escherichia coli topoisomerase IV. Purification, characterization, subunit structure, and subunit interactions. J Biol Chem 268:24481–24490
Reece RJ, Maxwell A (1991) DNA gyrase: structure and function. Crit Rev Biochem Mol 26:335–375
Levine C, Hiasa H, Marians KJ (1998) DNA gyrase and topoisomerase IV: biochemical activities, physiological roles during the chromosome replication, and drug sensitivities. Biochim Biophys Acta 1400:29–43
Ostrov DA, Hernández Prada JA, Corsino PE, Finton KA, Le N, Rowe TC (2007) Discovery of novel DNA gyrase inhibitors by high-throughput virtual screening. Antimicrob Agents Chemother 51:3688–3698
Zhang Y (2005) The magic bullets and tuberculosis drug targets. Annu Rev Pharmacol Toxicol 45:529–564
Vashist J, Vishvanath KR, Kapil A, Yennamalli Y, Subbarao N, Rajeswari MR (2009) Interaction of nalidixic acid and ciprofloxacin with wild type and mutated quinolone-resistance-determining region of DNA gyrase A. Indian J Biochem Biophys 46:147–153
Hooper DC (1999) Mode of action of fluoroquinolones. Drugs 58(suppl 2):6–10
Peterson LR (2001) Quinolone molecular structure-activity relationships: what we have learned about improving antimicrobial activity. Clin Infect Dis 33(Suppl 3):S180–S186
Ebalunode JO, Zheng W, Tropsha A (2011) Application of QSAR and shape pharmacophore modeling approaches for targeted chemical library design. Methods Mol Biol 685:111–133
Wolber G, Langer T (2005) LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J Chem Inf Comput Sci 45:160–169
Sippl W (2008) Pharmacophore identification and pseudo-receptor modeling. In: Wermuth CG (ed) The Practice of Medicinal Chemistry, 3rd edn. Academic Press/Elsevier, Amsterdam, pp 572–586
Perdih A, Kovač A, Wolber G, Blanot D, Gobec S, Solmajer T (2009) Discovery of novel benzene 1,3-dicarboxylic acid inhibitors of bacterial MurD and MurE ligases by structure-based virtual screening approach. Bioorg Med Chem Lett 19:2668–2673
Minovski N, Vračko M, Šolmajer T (2010) Quantitative structure-activity relationship study of antitubercular fluoroquinolones. Mol Div 15:417–426
Schwalbe T, Kadzimirisz D, Jas G (2000) Synthesis of a library of ciprofloxacin analogues by means of sequential organic synthesis in microreactors. QSAR Comb Sci 24:758–768
Martel AM, Leeson PA, Castañer J (1997) BAY-12–8039: fluoroquinolone antibacterial. Drugs Fut 22:109–113
Serradell MN, Blancafort P, Castañer J (1983) DL-8280. Drugs Fut 8:395
Key Organics, Bionet Fragment Library “Rule of 3”, http://www.keyorganics.ltd.uk
Congreve M, Carr R, Murray CW, Jhoti H (2003) A rule of three for fragment-based lead discovery? Drug Discovery Today 8:876–877
Minovski N, Šolmajer T (2010) Chemometrical exploration of combinatorially generated drug-like space of 6-fluoroquinolone analogs: a QSAR study. Acta Chim Slov 57:529–591
Wieland T (1997) Combinatorics of combinatorial chemistry. J Math Chem 21:141–157
Bohacek RS, mcMartin C, Guida WC (1996) The art and practice of structure-based drug design. Med Res Rev 16:3–50
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 23:3–25
Veber DF, Johnson SR, Cheng H-Y, Smith BR, Ward KW, Kopple KD (2002) Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 45:2615–2623
Katritzky AR, Lobanov VS, Karelson M (1995) CODESSA. Reference manual, University of Florida, Gainsville
Tretter EM, Schoeffler AJ, Weisfield SR, Berger JM (2010) Protein Struct Funct Bioinf 78:492–495
Fu G, Wu J, Liu W, Zhu D, Hu Y, Deng J, En Zhang X, Bi L, Cheng Wang D (2009) Crystal structure of DNA gyrase B′ domain sheds lights on the mechanism for T-segment navigation. Nucleic Acids Res 37:5908–5916
Laponogov I, Sohi MK, Veselkov DA, Pan XS, Sawhney R, Thompson AW, McAuley KE, Fisher LM, Sanderson MR (2009) Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat Struct Mol Biol 16:667–669
Kitamura A, Hoshino K, Kimura Y, Hayakawa I, Sato K (1995) Contribution of the C-8 substituent of DU-6859a, a new potent fluoroquinolone, to its activity against DNA gyrase mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemoter 39:1467–1471
Laponogov I, Pan XS, Veselkov DA, McAuley KE, Fisher LM, Sanderson MR (2010) Structural basis of gate-DNA breakage and resealing by type II topoisomerases. PloS One 5:e11338(1–8)
Jones G, Willet P, Glen RC, Leach AR, Taylor R (1997) Development and validation of a genetic algorithm for flexible docking. J Mol Biol 267:727–748
Kirchmair J, Markt P, Distinto S, Wolber G, Langer T (2008) Evaluation of the performance of 3D virtual screening protocols: RMSD comparisons, enrichment assessments, and decoy selection-What can we learn from earlier mistakes? J Comput Aided Mol Des 22:213–228
Huc I, Lehn J-M (1997) Virtual combinatorial libraries: Dynamic generation of molecular and supramolecular diversity by self-assembly. Proc Natl Acad Sci 94:2106–2110
Oprea TI, Gottfries J, Sherbukhin V, Svensson P, Kühler TC (2000) Chemical information management in drug discovery: Optimizing the computational and combinatorial chemistry interfaces. J Mol Graph Model 18:512–524
Langer T, Wolber G (2004) Virtual combinatorial chemistry and in silico screening: Efficient tools for lead structure discovery? Pur Appl Chem 76:991–996
Seneci P, Miertus S (2000) Combinatorial chemistry and high-throughput screening in drug discovery: different strategies and formats. Mol Diversity 5:75–89
Oprea TI (2002) Chemical space navigation in lead discovery. Curr Opin Chem Biol 6:384–389
Oprea TI (2000) Property distribution of drug-related chemical databases. J Comput Aided Mol Des 14:251–264
Allu TK, Oprea TI (2005) Rapid evaluation of synthetic and molecular complexity for in silico chemistry. J Chem Inf Model 45:1237–1243
Hann MM, Leach AR, Harper G (2001) Molecular complexity and its impact on the probability of finding leads for drug discovery. J Chem Inf Comput Sci 41:856–864
Madurga S, Sánchez-Céspedes J, Belda I, Vila J, Giralt E (2008) Mechanism of binding of fluoroquinolones to the quinolone resistance determining region of DNA gyrase: towards an understanding of the molecular basis of quinolone resistance. Chem Bio Chem 9:2081–2086
Acknowledgments
Authors thank Agency of Research of R. Slovenia (ARRS) for the financial support through the Grants P1-0017 and 1000-07-310016. We are sincerely grateful to Dr. Marjana Novič for valuable insights, discussion and her continuing support of this research.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online resource 1 (part 1)
The general virtual combinatorial library (CombiTot) of total 53.871 6-FQ structural analogs obtained using the virtual combinatorial chemistry approach (available as *.sdf file format). (TXT 83.0 MB)
Online resource 1 (part 2)
(TXT 50.0 MB)
Online resource 2
Chemical structures of the combinatorially-generated, filtered 6-FQ structural analogs marked as drug-like compounds (CombiDL, 1.101 compounds), obtained after the drug-likeness assessment (available as *.sdf file format). (TXT 3.74 MB)
Online resource 3
Chemical structures of the experimentally-determined 6-FQ structural analogs (Assay2, 145compounds), together with their measured biological activity values (MIC) (available as *.sdf file format). (TXT 476 KB)
Online resource 4
Chemical structures of the combinatorially-generated 6-FQ structural analogs (CombiLib, 427 compounds) extracted from the pool of 1.101 drug-like compounds (CombiDL), defining the global hypothetical activity range (GHA) of 0.0 ≤ MIC pred-combi [μg/mL] ≤ 0.1 (available as *.sdf file format). (TXT 1.41 MB)
Online resource 5
The experimental and combinatorial compounds, extracted as most promising 6-FQs (marked as true (T)) after the Boolean-type VS analysis using three-dimensional pharmacophore models (LBP, SBP shared , and SBP merged ). (XLS 205 KB)
Online resource 6
The post-docking first level VS analysis of both the sets (the experimental and the combinatorial one) into the 3K9F binding pocket. The geometrically-suitable compounds (with (T) outcome) are highlighted in green. (XLS 788 KB)
Online Resource 7
The post-docking second level VS analysis of the retained compounds from the first level post-docking analysis (experimental and combinatorial) into the 3K9F binding pocket. The selected compounds with optimal interatomic distances (with (T) outcome) are highlighted in green. (XLS 2.06 MB)
Rights and permissions
About this article
Cite this article
Minovski, N., Perdih, A. & Solmajer, T. Combinatorially-generated library of 6-fluoroquinolone analogs as potential novel antitubercular agents: a chemometric and molecular modeling assessment. J Mol Model 18, 1735–1753 (2012). https://doi.org/10.1007/s00894-011-1179-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-011-1179-0