Abstract
An understanding of why adenine (A) pairs with thymine (T) and cytosine (C) with guanine (G) in DNA is very useful in the design of sensors and other related devices. We report the use of dissociation energies, geometries and molecular electrostatic potentials (MEPs) to justify the canonical (AT and CG) Watson-Crick pairs. We also analyze all mismatches in both configurations—cis and trans—with respect to their glycoside bonds. As expected, we found that the most stable pair configuration corresponds to CG, providing an energy criterion for that preferred configuration. The reason why A gets together with T is much more difficult to explain as the energy of this pair is smaller than the energy of some other mismatched pairs. We tested MEPs to see if they could shed light on this problem. Interestingly, MEPs yield a unique pattern (shape) for the two canonical cases but different shapes for the mismatches. A tunnel of positive potential surrounded by a negative one is found interconnecting the three H-bonds of CG and the two of AT. This MEP tunnel, assisted partially by energetics and geometrical criteria, unambiguously determine a distinctive feature of the affinity between A and T as well as that between G and C.

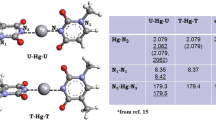
The signature of pairing in DNA: A characteristic positive potential tunnel on a negative background is observed in DNA; this potential shape (left) is an indicator of why A pairs with T and C pairs with G, as this shape form is not observed in most of others H-bonds




Similar content being viewed by others
References
Watson JD, Crick FHC (1953) Molecular structure of nucleic acids—a structure for deoxyribose nucleic acid. Nature 171:737–738
Kornberg A (1980) DNA replication. Freeman, San Francisco
Nir E, Plützer C, Kleinermanns K, de Vries M (2002) Properties of isolated DNA bases, base pairs and nucleosides examined by laser spectroscopy. Eur Phys J D 20:317–329
Showalter AK, Tsai MD (2001) A DNA polymerase with specificity for five base pairs. J Am Chem Soc 123:1776–1777. doi:10.1021/ja005758x
Friedberg EC, Walker GC, Siebe W (1995) DNA repair and mutagenesis. ASM, Washington, DC
Loft S, Poulsen HE (1996) Cancer risk and oxidative DNA damage in man. J Mol Spectrosc Med 74:297–312
Nir E, Kleinermanns K, de Vries MS (2000) Pairing of isolated nucleic-acid bases in the absence of the DNA backbone. Nature 408:949–951
Krueger AT, Kool ET (2007) Model systems for understanding DNA base pairing. Curr Opin Chem Biol 11:588–594
Edirisinghe N, Apalkov V, Berashevich J, Chakraborty T (2010) Electrical current through DNA containing mismatched base pairs. Nanotechnology 21:245101
Apalkov V, Berashevich J, Chakraborty T (2010) Unique magnetic signatures of mismatched base pairs in DNA. J Chem Phys 132:085102
Aboul-ela F, Koh D, Tinoco IJ (1985) Base-base mismatches. Thermodynamics of double helix formation for dCA3XA3G + dCT3YT3G (X, Y = A, C, G, T). Nucleic Acids Res 13:4811–4824
Schofield MJ, Hsieh P (2003) DNA MISMATCH REPAIR: Molecular mechanisms and biological function. Annu Rev Microbiol 57:579–608. doi:10.1146/annurev.micro.57.030502.090847
Modrich P (1987) DNA Mismatch correction. Annu Rev Biochem 56:435–466. doi:10.1146/annurev.bi.56.070187.002251
Werntges H, Steger G, Riesner D, Fritz HJ (1986) Mismatches in DNA double strands: thermodynamic parameters and their correlation to repair efficiencies. Nucleic Acids Res 14:3773–3790. doi:10.1093/nar/14.9.3773
Marra G, Schär P (1999) Recognition of DNA alterations by the mismatch repair system. Biochem J 338:1–13
Patel DJ, Ikuta S, Kozlowski S, Itakura K (1983) Sequence dependence of hydrogen exchange kinetics in DNA duplexes at the individual base pair level in solution. Proc Natl Acad Sci USA 80:2184–2188
Goodman MF (1997) Hydrogen bonding revisited: Geometric selection as a principal determinant of DNA replication fidelity. Proc Natl Acad Sci USA 94:10493–10495
Jauregui LA, Seminario JM (2008) A DNA sensor for sequencing and mismatches based on electron transport through Watson–Crick and Non-Watson–Crick base pairs. IEEE Sensors J 8:803–814
Moore CL, Zivkovic A, Engels JW, Kuchta RD (2004) Human DNA primase uses Watson-Crick hydrogen bonds to distinguish between correct and incorrect nucleoside triphosphates. Biochemistry 43:12367–12374. doi:10.1021/bi0490791
Wolfle WT, Washington MT, Kool ET, Spratt TE, Helquist SA, Prakash L, Prakash S (2005) Evidence for a Watson-Crick hydrogen bonding requirement in DNA synthesis by human DNA polymerase {kappa}. Mol Cell Biol 25:7137–7143. doi:10.1128/mcb.25.16.7137-7143.2005
Mizukami S, Kim TW, Helquist SA, Kool ET (2006) Varying DNA base-pair size in subangstrom increments: evidence for a loose, not large, active site in low-fidelity Dpo4 polymerase. Biochemistry 45:2772–2778. doi:10.1021/bi051961z
Kim TW, Delaney JC, Essigmann JM, Kool ET (2005) Probing the active site tightness of DNA polymerase in subangstrom increments. Proc Natl Acad Sci USA 102:15803–15808. doi:10.1073/pnas.0505113102
Kim TW, Brieba LG, Ellenberger T, Kool ET (2006) Functional evidence for a small and rigid active site in a high fidelity DNA polymerase. J Biol Chem 281:2289–2295. doi:10.1074/jbc.M510744200
Mitsui T, Kimoto M, Harada Y, Yokoyama S, Hirao I (2005) An efficient unnatural base pair for a base-pair-expanded transcription system. J Am Chem Soc 127:8652–8658. doi:10.1021/ja0425280
Yang Z, Sismour AM, Sheng P, Puskar NL, Benner SA (2007) Enzymatic incorporation of a third nucleobase pair. Nucleic Acids Res 35:4238–4249. doi:10.1093/nar/gkm395
Sismour AM, Benner SA (2005) The use of thymidine analogs to improve the replication of an extra DNA base pair: a synthetic biological system. Nucleic Acids Res 33:5640–5646. doi:10.1093/nar/gki873
Danilov VI, Anisimov VM (2005) Post Hartree-Fock studies of the canonical Watson-Crick DNA base pairs: molecular structure and the nature of stability. J Biomol Struct Dyn 22:471–482
Hobza P, Sponer J (1999) Structure, energetics, and dynamics of the nucleic acid base pairs: nonempirical ab initio calculations. Chem Rev 99:3247–3276
Guerra CF, Bickelhaupt FM, Snijders JG, Baerends EJ (2000) Hydrogen bonding in DNA base pairs: reconciliation of theory and experiment. J Am Chem Soc 122:4117–4128
Das J, Mukherjee S, Mitra A, Bhattacharyya D (2006) Non-canonical base pairs and higher order structures in nucleic acids: crystal structure database analysis. J Biomol Struct Dyn 24:149–161
Bhattacharyya D, Koripella SC, Mitra A, Rajendran VB, Sinha B (2007) Theoretical analysis of noncanonical base pairing interactions in RNA molecules. J Biosci 32:809–825
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA (2004) Gaussian 03, Revision C.02. Gaussian Inc, Wallingford CT
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JA Jr, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam JM, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas O, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ (2009) Gaussian 09, Revision A.02. Gaussian Inc, Wallingford CT
Lescoute A, Westhof E (2006) The interaction networks of structured RNAs. Nucleic Acids Res 34:6587–6604
Leontis NB, Westhof E (1998) Conserved geometrical base-pairing patterns in RNA. Q Rev Biophys 31:399–455. doi:10.1017/S0033583599003479
Leontis NB, Westhof E (2001) Geometric nomenclature and classification of RNA base pairs. RNA 7:499–512
Becke AD (1993) A new mixing of Hartree–Fock and local density-functional theories. J Chem Phys 98:1372–1377
Perdew JP (1991) Unified theory of exchange and correlation beyond the local density approximation. In: Ziesche P, Eschrig H (eds) Electronic structure of solids. Akademie, Berlin, pp 11–20
Petersson GA, Bennett A, Tensfeldt TG, Al-Laham MA, Shirley WA, Mantzaris J (1988) A complete basis set model chemistry. I. The total energies of closed-shell atoms and hydrides of the first-row elements. J Chem Phys 89:2193–2218
Petersson GA, Al-Laham MA (1991) A complete basis set model chemistry. II. Open-shell systems and the total energies of the first-row atoms. J Chem Phys 94:6081–6090
Habibollahzadeh D, Grodzicki M, Seminario JM, Politzer P (1991) Computational study of the concerted gas-phase triple dissociations of 1,3,5-triazacyclohexane and its 1,3,5-trinitro derivative (RDX). J Phys Chem 95:7699–7702
Politzer P, Seminario JM (1993) Computational study of the structure of dinitraminic acid, HN(NO2)2 and the energetics of some possible decomposition steps. Chem Phys Lett 216:348–352
Seminario JM, Concha MC, Politzer P (1995) A density functional/molecular dynamics of the structure of liquid nitromethane. J Chem Phys 102:8281–8282
Politzer P, Seminario JM (1993) Energy changes associated with some decomposition steps of 1,3,3-trinitroazetidine—a nonlocal density-functional study. Chem Phys Lett 207:27–30
Politzer P, Seminario JM, Bolduc PR (1989) A proposed interpretation of the destabilizing effect of hydroxyl-groups on nitroaromatic molecules. Chem Phys Lett 158:463–469
Seminario JM, Concha MC, Politzer P (1992) Calculated sructures and relative tabilities of furoxan, some 1,2 dinitrosoethylenes and other isomers. J Comput Chem 13:177–182
Murray JS, Redfern PC, Seminario JM, Politzer P (1990) Anomalous energy effects in some aliphatic and alicyclic aza systems and their nitro-derivatives. J Phys Chem A 94:2320–2323
Seminario JM, Araujo RA, Yan L (2004) Negative differential resistance in metallic and semiconducting clusters. J Phys Chem B 108:6915–6918
Seminario JM, De La Cruz C, Derosa PA, Yan L (2004) Nanometer-size conducting and insulating molecular devices. J Phys Chem B 108:17879–17885
Derosa PA, Guda S, Seminario JM (2003) A programmable molecular diode driven by charge-induced conformational changes. J Am Chem Soc 125:14240–14241
Seminario JM, Zacarias AG, Castro M (1997) Systematic study of the lowest energy states of Pd, Pd2, and Pd3. Int J Quantum Chem 61:515–523
Seminario JM, Tour JM (1997) Systematic study of the lowest energy states of Aun (n = 1–4) Using DFT. Int J Quantum Chem 65:749–758
Derosa PA, Seminario JM, Balbuena PB (2001) Properties of small bimetallic Ni-Cu clusters. J Phys Chem A 105:7917–7925
Seminario JM, Ma Y, Agapito LA, Yan L, Araujo RA, Bingi S, Vadlamani NS, Chagarlamudi K, Sudarshan TS, Myrick ML, Colavita PE, Franzon PD, Nackashi DP, Cheng L, Yao Y, Tour JM (2004) Clustering effects on discontinuous gold film nanocells. J Nanosci Nanotechnol 4:907–917
Seminario JM, Derosa PA, Cordova LE, Bozard BH (2004) A molecular device operating at Terahertz frequencies. IEEE Trans Nanotech 3:215–218
Choi D-S, Huang S, Huang M, Barnard TS, Adams RD, Seminario JM, Tour JM (1998) Revised structures of N-substituted dibrominated pyrrole derivatives and their polymeric products. Termaleimide models with low optical bandgaps. J Org Chem 63:2646–2655
Murray JS, Seminario JM, Politzer P (1994) Does antiaromaticity imply net destabilization. Int J Quantum Chem 49:575–579
Politzer P, Seminario JM (1989) Computational analysis of the structures, bond properties, and electrostatic potentials of some nitrotetrahedranes and nitroazatetrahedranes. J Phys Chem 93:4742–4745
Seminario JM, Politzer P (1991) First principles theoretical methods for the calculation of electronic charge densities and electrostatic potentials. In: Jeffrey GA (ed) The application of charge-density research to chemistry and drug design. Plenum, New York
Politzer P, Grice ME, Murray JS, Seminario JM (1993) Anomalous stabilizing and destabilizing effects in some cyclic pi-electron systems. Can J Chem 71:1123–1127
Seminario JM, Concha MC, Murray JS, Politzer P (1994) Theoretical analyses of O2/H2O systems under normal and supercritical conditions. Chem Phys Lett 222:25–32
Hohenberg P, Kohn W (1964) Inhomogeneous electron gas. Phys Rev 136:B864
Kohn W, Sham LJ (1965) Self-consistent equations including exchange and correlation effects. Phys Rev 140:A1133
Sham LJ, Kohn W (1966) One-particle properties of an inhomogeneous interacting electron gas. Phys Rev 145:561–567
Murray JS, Peralta-Inga Z, Politzer P, Ekanayake K, LeBreton P (2001) Computational characterization of nucleotide bases: Molecular surface electrostatic potentials and local ionization energies, and local polarization energies. Int J Quantum Chem 83:245–254
Politzer P, Seminario JM (1990) Relative bond strengths in tetrahedrane, prismane, and some of their aza analogs. Struct Chem 1:29–32
Scrocco E, Tomasi J (1973) The electrostatic molecular potential as a tool for the interpretation of molecular properties. In: New concepts II, vol 42. Topics in current chemistry. Springer. Berlin, pp 95–170. doi:10.1007/3-540-06399-4_6
Jauregui LA, Salazar-Salinas K, Seminario JM (2009) Transverse electronic transport in double-stranded DNA nucleotides. J Phys Chem B 113:6230–6239. doi:10.1021/jp808790j
Sivanesan D, Subramanian V, Nair BU (2001) Solvent effect on DNA base stacked dimers: an isodensity polarizable continuum model approach. Int J Quantum Chem 84:750–758
Marañon J, Fantoni A, Grigera JR (1999) Adenine-thymine molecular dynamics simulation. Conformation, hydration and magnetic behaviour. J Mol Liq 79:177–186
Sponer J, Sabat M, Burda JV, Leszczynski J, Hobza P (1999) Interaction of the adenine–thymine Watson –Crick and adenine–adenine reverse-Hoogsteen DNA base pairs with hydrated group IIa (Mg2+, Ca2+, Sr2+, Ba2+) and IIb (Zn2+, Cd2+, Hg2+) metal cations: absence of the base pair stabilization by metal-induced polarization effects. J Phys Chem B 103:2528–2534. doi:10.1021/jp983744w
Coutinho K, Ludwig V, Canuto S (2004) Combined Monte Carlo and quantum mechanics study of the hydration of the guanine-cytosine base pair. Phys Rev E 69:061902
Gilbert HF (2000) Basic concepts in biochemistry, 2nd edn. McGraw-Hill, Houston
Tikhomirova A, Beletskaya IV, Chalikian TV (2006) Stability of DNA duplexes containing GG, CC, AA, and TT mismatches. Biochemistry 45:10563–10571. doi:10.1021/bi060304j
Hunter WN, Brown T, Anand NN, Kennard O (1986) Structure of an adenine[dot]cytosine base pair in DNA and its implications for mismatch repair. Nature 320:552–555
Acknowledgments
We thank the Defense Threat Reduction Agency (DTRA) through the Army Research Office (ARO) for their support, Project No. W911NF-06-1-0231 and the ARO for the DURIP/ARO Project # W91NF-07-1-0199 and the MURI/ARO Project # W911NF-11-1-0024.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Otero-Navas, I., Seminario, J.M. Molecular electrostatic potentials of DNA base–base pairing and mispairing. J Mol Model 18, 91–101 (2012). https://doi.org/10.1007/s00894-011-1028-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-011-1028-1