Abstract
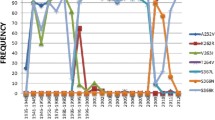
As one of the most important antiviral drugs against 2009 influenza A (H1N1), will zanamivir be effective for the possible drug resistant mutants? To answer this question, we combined multiple molecular dynamics simulations and molecular mechanics generalized Born surface area (MM-GBSA) calculations to study the efficiency of zanamivir over the most frequent drug-resistant strains of neuraminidase including R293K, R152K, E119A/D and H275Y mutants. The calculated results indicate that the modeled mutants of the 2009-H1N1 strains except H275Y will be significantly resistant to zanamivir. The resistance to zanamivir is mainly caused by the loss of polar interactions. The identified potential resistance sites in this study will be useful for the development of new effective anti-influenza drugs and to avoid the occurrence of the state without effective drugs to new mutant influenza strains.

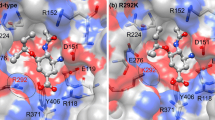
The studied mutations of neuraminidase and their influence to zanamivir binding






Similar content being viewed by others
References
Garten RJ, Davis CT, Russell CA et al (2009) Antigenic and genetic characteristics of swine-origin 2009 A (H1N1) influenza viruses circulating in humans. Science 325:197–201
CDC (2010) Antiviral Drugs and H1N1 Flu (Swine Flu). http://www.cdc.gov/h1n1flu/antiviral.htm. Accessed 10 July 2010)
Abed Y, Nehmé B, Baz M, Boivin G (2008) Activity of the neuraminidase inhibitor A-315675 against oseltamivir-resistant influenza neuraminidases of N1 and N2 subtypes. Antivir Res 77:163–166
Collins PJ, Haire LF, Lin YP, Liu J, Russell RJ, Walker PA, Skehel JJ, Martin SR, Hay AJ, Gamblin SJ (2008) Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants. Nature 453:1258–1261
Boivin G, Goyette N (2002) Susceptibility of recent Canadian influenza A and B virus isolates to different neuraminidase inhibitors. Antivir Res 54:143–147
Zürcher T, Yates PJ, Daly J, Sahasrabudhe A, Walters M, Dash L, Tisdale M, McKimm-Breschkin JL (2006) Mutations conferring zanamivir resistance in human influenza virus N2 neuraminidases compromise virus fitness and are not stably maintained in vitro. J Antimicrob Chemoth 58:723–732
Mishin VP, Hayden FG, Gubareva LV (2005) Susceptibilities of antiviral-resistant influenza viruses to novel neuraminidase inhibitors. Antimicrob Agents Chemoth 49:4515–4520
Arias CF, Escalera-Zamudio M, Soto-Del Río Mde L, Cobián-Güemes AG, Isa P, López S (2009) Molecular Anatomy of 2009 Influenza Virus A (H1N1). Arch Med Res 40:643–654
Gubareva LV, Webster RG, Hayden FG (2002) Detection of influenza virus resistance to neuraminidase inhibitors by an enzyme inhibition assay. Antivir Res 53:47–61
http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5832a3.htm (accessed August 9, 2010)
Cao ZW, Han LY, Zheng CJ, Ji ZL, Chen X, Lin HH, Chen YZ (2005) Computer prediction of drug resistance mutations in proteins. Drug Discov Today 10:521–529
Chachra R, Rizzo RC (2008) Origins of resistance conferred by the R292K neuraminidase mutation via molecular dynamics and free energy calculations. J Chem Theor Comput 4:1526–1540
Hou T, McLaughlin WA, Wang W (2008) Evaluating the potency of HIV-1 protease drugs to combat resistance. Proteins 71:1163–1174
Zhang J, Hou T, Wang W, Liu JS (2010) Detecting and understanding combinatorial mutation patterns responsible for HIV drug resistance. Proc Natl Acad Sci USA 107:1321–1326
Srinivasan J, Cheatham TE, Cieplak P, Kollman PA, Case DA (1998) Continuum solvent studies of the stability of DNA, RNA, and phosphoramidate–DNA helices. J Am Chem Soc 120:9401–9409
Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE (2000) Calculating structures and free energies of complex molecules: combining molecular Molecular mechanics and continuum models. Acc Chem Res 33:889–897
Tsui V, Case DA (2000) Theory and applications of the generalized born solvation model in macromolecular simulations. Biopolymers 56:275–291
Onufriev A, Bashford D, Case DA (2000) Modification of the generalized Born model suitable for macromolecules. J Phys Chem B 104:3712–3720
Feig M, Brooks CL (2002) Evaluating CASP4 predictions with physical energy functions. Proteins 49:232–245
Gourmala C, Luo Y, Barbault F, Zhang Y, Ghalem S, Maurel F, Fan B (2007) Elucidation of the LewisX–LewisX carbohydrate interaction with molecular dynamics simulations: A glycosynapse model. J Mol Struct THEOCHEM 821:22–29
Shaikh SA, Jayaram B (2007) A swift all-atom energy-based computational protocol to predict DNA-ligand binding affinity and △Tm. J Med Chem 50:2240–2244
Liu H, Yao X, Wang C, Han J (2010) In silico identification of the potential drug resistance sites over 2009 Influenza A (H1N1) virus neuraminidase. Mol Pharmaceutics 7:894–904
Rungrotmongkol T, Malaisree M, Nunthaboot N, Sompornpisut P, Hannongbua S (2010) Molecular prediction of oseltamivir efficiency against probable influenza A (H1N1-2009) mutants: molecular modeling approach. Amino Acids 39:393–398
DeLano WL (2002) The PyMOL Molecular Graphics System DeLano Scientific, Palo Alto, CA, USA. http://www.pymol.org
Li Q, Qi J, Zhang W, Vavricka CJ, Shi Y, Wei J, Feng E, Shen J, Chen J, Liu D, He J, Yan J, Liu H, Jiang H, Teng M, Li X, Gao GF (2010) The 2009 pandemic H1N1 neuraminidase N1 lacks the 150-cavity in its active site. Nat Struct Mol Biol 17:1266–1268
Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ (2005) The Amber biomolecular simulation programs. J Comput Chem 26:1668–1688
Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman PA (2003) A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem 24:1999–2012
Lee MC, Duan Y (2004) Distinguish protein decoys by using a scoring function based on a new AMBER force field, short molecular dynamics simulations, and the generalized born solvent model. Proteins: Struct Funct Bioinform 55:620–634
Wang JM, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general amber force field. J Comput Chem 25:1157–1174
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935
Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690
Essmann U, Perera L, Berkowitz ML, Darden TA (1995) Smooth particle mesh Ewald method. J Chem Phys 103:8577–9593
Ryckaert JP, Ciccotti G, Berendsen HJC (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23:327–341
Sitkoff D, Sharp KA, Honig B (1994) Accurate calculation of hydration free energies using macroscopic solvent models. J Phys Chem 98:1978–1988
Case DA (1994) Normal mode analysis of protein dynamics. Curr Opin Struct Biol 4:285–290
Varghese JN, Smith PW, Sollis SL (1998) Drug design against a shifting target: a structural basis for resistance to inhibitors in a variant of influenza virus neuraminidase. Structure 6:735–746
Smith BJ, McKimm-Breshkin JL, McDonald M (2002) Structural studies of the resistance of influenza virus neuramindase to inhibitors. J Med Chem 45:2207–2212
McKimm-Breschkin J, Sahasrabudhe A, Blick T (2001) Mechanisms of resistance of influenza virus to neuraminidase inhibitors. Int Congress Series 1219:855–861
Chang CA, Chen W, Gilson MK (2007) Ligand configurational entropy and protein binding. Proc Natl Acad Sci USA 104:1534–1539
Gohlke H, Klebe G (2002) Approaches to the description and prediction of the binding affinity of small-molecule ligands to macromolecular receptors. Angew Chem Int Edn 41:2644–2676
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No: 20905033) and the Fundamental Research Funds for the Central Universities (Grant No: lzujbky-2009-97). The authors also would like to thank the Gansu Computing Center for providing the computing resources.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pan, D., Sun, H., Bai, C. et al. Prediction of zanamivir efficiency over the possible 2009 Influenza A (H1N1) mutants by multiple molecular dynamics simulations and free energy calculations. J Mol Model 17, 2465–2473 (2011). https://doi.org/10.1007/s00894-010-0929-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-010-0929-8