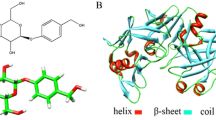
Abstract
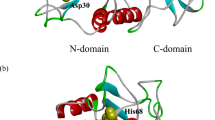
In the structural-based mutagenesis of Mucor pusillus pepsin (MPP), understanding how κ-casein interacts with MPP is a great challenge for us to explore. Chymosin-sensitive peptide is the key domain of κ-casein that regulates milk clotting through the specific proteolytic cleavage of its peptide bond (Phe105-Met106) by MPP to produce insoluble para-κ-casein. Here, we built the model of this large peptide using molecular modeling technique. Docking study showed that MPP can accommodate the designed model with a favorable binding energy and the docked complex has proven to locally resemble the inhibitor-chymosin complex. The catalytic mechanism for the peptide model binding with MPP was explored in terms of substrate-enzyme interaction and property of contacting surface. Some critical amino acid residues in the substrate binding cleft have been identified as an important guide for further site-directed mutagenesis. Glu13 and Leu11 in the S3 region of MPP, predicted as the special mutation sites, were confirmed to retain clotting activity and decrease the proteolytic activity. These novel mutants may provide a promising application for improving cheese flavor.

Novel mutants of mucor pusillus pepsin having a promising application for improving cheese flavor were found by using molecular modeling technology.






Similar content being viewed by others
References
Beppu T (1983) Trends Biotechnol 1:85–89
Foltman B (1966) CR Trav Lab Carlsberg 35:143–231
Drøhse HB, Foltmann B (1989) Biochim Biophys Acta 995:221–224
Martin P, Raymond MN, Bricas E, Dumas BR (1980) Biochim Biophys Acta 612:410–420
Jollès P, Alais C, Jollès J (1963) Biochim Biophys Acta 69:511–517
Egitoa AS, Girardetc JM, Lagunaa LE, Poirsonc C, Molléb D, Micloc L, Humbertc G, Gaillardc JL (2007) Int Dairy J 17:816–825
Yamashita T, Higashi S, Higashi T, Machida H, Iwasaki S, Nishiyama M, Beppu T (1994) J Biotechnol 32:17–28
Branner-Jørgensen S, Eigtved P, Schneider P (1981) Neth Milk Dairy J 35:361–364
Yamashita T, Tonouchi N, Uozumi T, Beppu T (1987) Mol Gen Genet 210:462–467
Hiramatsu R, Aikawa J, Horinouchi S, Beppu T (1989) J Biol Chem 264:16862–16866
Hiramatsu R, Yamashita T, Aikawa J, Horinouchi S, Beppu T (1990) Appl Environ Microbiol 56:2125–2132
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Nucleic Acids Res 25:3389–3402
Ouali M, King RD (2000) Protein Sci 9:1162–1176
Pollastri G, Baldi P (2002) Bioinformatics 18(Suppl 1):S62–S70
Lin K, Simossis VA, Taylor WR, Heringa J (2005) Bioinformatics 21:152–159
Jones DT (1999) J Mol Biol 292:195–202
Simossis VA, Heringa J (2004) Bioinformatics (in press)
InsightII, Homology User Guide, SanDiego:Biosym/MSI (2000)
Newman M, Watson F, Roychowdhury P, Jones H, Badasso M, Cleasby A, Wood SP, Tickle IJ, Blundell TL (1993) J Mol Biol 230:260–283
Affinity San Diego Molecular Simulations Inc (2000)
Bartlett PA, Shea GT, Telfer SJ, Waterman S (1989) R Soc Chem 182–196
Shoichet BK, Kuntz ID, Bodian DL (1992) J Comput Chem 13:380–397
Kunkel TA (1985) Proc Natl Acad Sci USA 82:488–492
Zhang J, Zhang SQ, Wu X, Chen YQ, Diao ZY (2006) Process Biochem 41:251–256
International Dairy Federation: Brussels, Belgium (1987) Calf rennet and adult bovine rennet:Determination of chymosin and bovine pepsin contents (chromatographic method). Standard 110A
Chitpinityol S, Goode D, Crabbe MJC (1998) Food Chem 62:133–139
Yamashita MM, Almassy RJ, Janson CA, Cascio D, Eisenberg D (1989) J Biol Chem 264:17681–17690
Groves MR, Dhanaraj V, Badasso M, Nugent P, Pitts JE, Hoover DJ, Blundell TL (1998) Protein Eng 11:833–840
Chitpinityol S, Crabbe MJC (1998) Food Chem 61:395–418
Pearl LH (1987) FEBS Lett 214:8–12
Park YN, Aikawa J, Nishiyama M, Horinouchi S, Beppu T (1996) Protein Eng 9:869–875
Aikawa J, Yamashita T, Nishiyama M, Horinouchi S, Beppu T (1990) J Biol Chem 265:13955–13959
Acknowledgments
This work was supported by National 863 Program 2006AA10Z306, National Public Benefit Research (Agriculture) Foundation (200903043), China Postdoctoral Science Foundation funded project (20100471246), Natural Science Foundation of China (31071574), Natural Science Foundation for the Youth (21004028) and The Earmarked Fund for Modern Agro-industrial Technology Research Systems in China (Nycytx-05-02).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 100 kb)
Rights and permissions
About this article
Cite this article
Li, T., Wang, J., Li, Y. et al. Stucture of the complex between Mucor pusillus pepsin and the key domain of κ-casein for site-directed mutagenesis: a combined molecular modeling and docking approach. J Mol Model 17, 1661–1668 (2011). https://doi.org/10.1007/s00894-010-0869-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-010-0869-3