Abstract
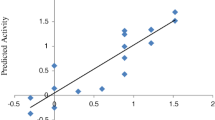
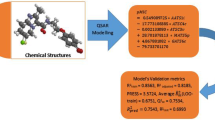
In the current study, the applicability and scope of descriptor based QSAR models to complement virtual screening using molecular docking approach have been applied to identify potential virtual screening hits targeting DNA gyrase A from Mycobacterium tuberculosis, an effective and validated anti-mycobacterial target. Initially QSAR models were developed against M. fortuitum and M. smegmatis using a series of structurally related fluoroquinolone derivatives as DNA gyrase inhibitors. Both the QSAR models yielded significant cross validated Q(2) values of 0.6715 and 0.6944 and R(2) values of 0.7250 and 0.7420, respectively. The statistically significant models were validated by a test set of 22 compounds with predictive R(2) value of 0.7562 and 0.7087 for M. fortuitum and M. smegmatis respectively. To aid the creation of novel antituberculosis compounds, combinatorial library was developed on fluoroquinolone template to derive a data set of 5280 compounds whose activity values have been measured by the above models. Highly active compounds predicted from the models were subjected to molecular docking study to investigate the mechanism of drug binding with the DNA gyrase A protein of M. tuberculosis and the compounds showing similar type of binding patterns with that of the existing drug molecules, like sparfloxacin, were finally reported. It is seen that hydrophobic characteristics of molecular structure together with few hydrogen bond interactions are playing an essential role in antimicrobial activity for the fluoroquinolone derivatives. A representative set of seven compounds with high predicted MIC values were sorted out in the present study.







Similar content being viewed by others
References
Cambau E, Sougakoff W, Besson M, Truffot-Pernot C, Grosset J, Jarlier V (1994) Selection of a gyrA mutant of Mycobacterium tuberculosis resistant to fluoroquinolones during treatment with ofloxacin. J Infect Dis 170:479–483
Grosset JH (1992) Treatment of tuberculosis in HIV infection. Tuberc Lung Dis 73:378–383
Tsukamura M, Nakamura E, Yoshii S, Amano H (1985) Therapeutic effect of a new antibacterial substance ofloxacin (DL8280) on pulmonary tuberculosis. Am Rev Respir Dis 131:352–356
Blumberg HM, Burman WJ, Chaisson RE, Daley CL, Etkind SC et al. (2003) American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America: treatment of tuberculosis. Am J Respir Crit Care Med 167:603–662
Crofton J, Chaulet P, Maher D, Grosset J, Harris W et al. (1997) Guidelines for the management of drug-resistant tuberculosis. World Health Organization, Geneva, Switzerland
Takiff HE, Salazar L, Guerrero C, Philipp W, Huang WM et al. (1994) Cloning and nucleotide sequence of Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob Agents Chemother 38:773–780
Aubry A, Pan XS, Fisher LM, Jarlier V, Cambau E (2004) Mycobacterium tuberculosis DNA gyrase: interaction with quinolones and correlation with antimycobacterial drug activity. Antimicrob Agents Chemother 48:1281–1288
Maxwell A (1997) DNA gyrase as a drug target. Trends Microbiol 5:102–109
Willmott CJR, Maxwell A (1993) A single point mutation in the DNA gyrase A protein greatly reduces binding of fluoroquinolones to the gyrase-DNA complex. Antimicrob Agents Chemother 37:126–127
Barnard FM, Maxwell A (2001) Interaction between DNA Gyrase and Quinolones: effects of Alanine mutations at GyrA subunit residues Ser83 and Asp87. Antimicrob Agents Chemother 45:1994–2000
Hiasa H, Shea ME (2000) DNA gyrase-mediated wrapping of the DNA strand is required for the replication fork arrest by the DNA gyrase-quinolone-DNA ternary complex. J Biol Chem 275:34780–34786
Wentzell LM, Maxwell A (2000) The complex of DNA gyrase and quinolone drugs on DNA forms a barrier to the T7 DNA polymerase replication complex. J Mol Biol 304:779–791
Renau TE, Sanchiez JP, Gage JW, Dever JA, Shapiro MA et al. (1996) Structure-activity relationships of the Quinolone antibacterials against mycobacteria: effect of structural changes at N-1 and C-7. J Med Chem 39:729–735
Renau TE, Gage JW, Dever JA, Roland GE, Joannides ET et al. (1996) Structure-activity relationships of Quinolone agents against mycobacteria: effect of structural modifications at the 8 position. Antimicrob Agents Chemother 40:2363–2368
Bagchi MC, Maiti BC, Mills D, Basak SC (2004) Usefulness of graphical invariants in quantitative structure-activity correlations of tuberculostatic drugs of the isonicotinic acid hydrazide type. J Mol Model 10:102–111
Ghosh P, Thanadath M, Bagchi MC (2006) On an aspect of calculated molecular descriptors in QSAR studies of quinolone antibacterials. Mol Divers 10:415–27
Bagchi MC, Mills D, Basak SC (2007) Quantitative structure-activity relationship(QSAR) studies of antibacterials against M. fortuitum and M. smegmatis using theoretical molecular descriptors. J Mol Model 13:111–120
Ghosh P, Vracko M, Chattopadhyay AK, Bagchi MC (2008) On application of constitutional descriptors for merging of Quinoxaline data sets using linear statistical methods. Chem Biol Drug Design 72:155–162
Golbraikh A, Tropsha A (2002) Predictive QSAR modeling based on diversity sampling of experimental datasets for the training and test set selection. J Comput Aided Mol Des 16:357–369
Molecular Design Suite 3.5, VLife Technologies, Pune, India. www.vlifesciences.com
Guyon I, Elisseeff A (2003) An introduction to variable and feature selection. J Mach Learn Res 3:1157–1182
Hasegawa K, Kimura T, Funatsu K (1999) GA strategy for variable selection in QSAR studies:enhancement of comparative molecular binding energy analysis by GA-based PLS method. Quant Struct Act Relat 18:262–272
Ghosh P, Bagchi MC (2009) QSAR modeling for Quinoxaline derivatives using genetic algorithm and simulated annealing based feature selection. Curr Med Chem 16:4032–4048
Ghosh P, Bagchi MC (2009) Comparative QSAR studies of nitrofuranyl amide derivatives using theoretical structural properties. Mol Simulat 35:1185–1200
Holland JH (1992) Genetic algorithms. Sci Am 267:66–72
Luco JM, Ferretti FH (1997) QSAR based on multiple linear regression and PLS methods for the anti-HIV activity of a large group of HEPT derivatives. J Chem Inf Comput Sci 37:392–401
Leonard JT, Roy K (2006) QSAR by LFER model of HIV protease inhibitor mannitol derivatives using FA-MLR, PCRA, and PLS techniques. Bioorg Med Chem 14:1039–1046
Leach AR, Gillet VJ (2003) An Introduction to Chemoinformatics. Kluwer, Boston, pp 79–81
Martin YC (2001) Diverse viewpoints on computational aspects of molecular diversity. J Comb Chem 3:231–250
Böhm HJ, Stahl M (2000) Structure-based library design: molecular modeling merges with combinatorial chemistry. Curr Opin Chem Biol 4:283–286
Beavers MP, Chen Z (2002) Structure-based combinatorial library design: methodologies and applications. J Mol Graph Model 20:463–468
Hansch C, Leo A (1995) Exploring QSAR:Fundamentals and Applications in Chemistry and Biology. American Chemical Society, Washington, DC
Lajiness M (1990) Computational Chemical Graph Theory. Ed Rouvray, DH (Nova, New York), pp 299–316
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 23:3–25
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL Workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201
Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucl Acids Res 31:3381–3385
Laskowski RA, MacArthur MW, Moss D, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291
Gehlhaar DK, Verkhivker GM, Rejto PA, Sherman CJ, Fogel D et al. (1995) Molecular recognition of the inhibitor AC-1343 by HIV-l protease: conformationally flexible docking by evolutionary programming. Chem Biol 2:317–324
Verkhivker GM, Bouzida D, Gehlhaar DK, Rejto PA, Arthurs S et al. (2000) Deciphering common failures in molecular docking of ligand–protein complexes. J Comput Aided Mol Des 14:731–751
Ajmani S, Karanam S, Kulkarni SA (2009) Rationalizing protein-ligand interactions for PTP1B inhibitors using computational methods. Chem Biol Drug Des 74:582–595
Tretter EM, Schoeffler AJ, Weisfield SR, Berger JM (2010) Crystal structure of the DNA Gyrase GyrA N-terminal domain from Mycobacterium tuberculosis. Proteins 78:492–495
Madurga S, Sánchez-Céspedes J, Belda I, Vila J, Giralt E (2008) Mechanism of binding of fluoroquinolones to the quinolone resistance-determining region of DNA gyrase: towards an understanding of the molecular basis of quinolone resistance. Chem Bio Chem 9(13):2081–2086
Maxwell A (1992) The molecular basis of quinolone action. J Antimicrob Chemother 30:409–416
Tropsha A, Gramatica P, Gombar VK (2003) The importance of being earnest: validation is the absolute essential for successful application and interpretation of QSPR models. QSAR Comb Sci 22:69–77
Acknowledgments
Payel Ghosh thanks the Council of Scientific and Industrial Research, New Delhi 110001, India, for the grant of a Senior Research Fellowship to her. MCB acknowledges the Department of Biotechnology, New Delhi, India, for the grant of a project. The infrastructural support received from Bioinformatics Centre, IICB is also acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ghosh, P., Bagchi, M.C. Anti-tubercular drug designing by structure based screening of combinatorial libraries. J Mol Model 17, 1607–1620 (2011). https://doi.org/10.1007/s00894-010-0861-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-010-0861-y