Abstract
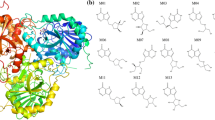
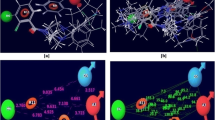
Recently, the massively parallel computation of absolute binding free energy with a well-equilibrated system (MP-CAFEE) has been developed. The present study aimed to determine whether the MP-CAFEE method is useful for drug discovery research. In the drug discovery process, it is important for computational chemists to predict the binding affinity accurately without detailed structural information for protein / ligand complex. We investigated the absolute binding free energies for Poly (ADP-ribose) polymerase-1 (PARP-1) / inhibitor complexes, using the MP-CAFEE method. Although each docking model was used as an input structure, it was found that the absolute binding free energies calculated by MP-CAFEE are well consistent with the experimental ones. The accuracy of this method is much higher than that using molecular mechanics Poisson-Boltzmann / surface area (MM / PBSA). Although the simulation time is quite extensive, the reliable predictor of binding free energies would be a useful tool for drug discovery projects.




Similar content being viewed by others
References
Nguewa PA, Fuertes MA, Alonso C, Petéz JM (2003) Mol Pharmacol 64:1007–1014
LaPlaca MC, Zhang J, Raghupathi R, Li JH, Smith F, Bareyre FM, Snyder SH, Graham DI, Mclntosh TK (2001) J Neurotrauma 18:369–376
Mandir AS, Przedborski S, Jackson-Lewis V, Wang Z, Simbulan-Rosenthal CM, Smulson ME, Hoffman BE, Guastella DB, Dawson VL, Dawson TM (1999) Proc Natl Acad Sci USA 96:5774–5779
Menear KA, Adcock C, Boulter R, Cockcroft XL, Cranston CL, Dillon KJ A, Drzewiecki J, Garman S, Gomez S, Javaid H, Kerrigan F, Knights C, Lau A, Loh VM Jr, Matthews IT, Moore S, O'Connor MJ, Smith GC, Martin NM (2008) J Med Chem 51:6581–6591
Southan GJ, Szabó C (2003) Curr Med Chem 10:321–340
Meirovitch H, Cheluvaraja S, White RP (2009) Curr Protein Pept Sci 10:229–243
Fukunishi Y, Mitomo D, Nakamura H (2009) J Chem Inf Model 49:1944–1951
Card GL, Blasdel L, England BP, Zhang C, Suzuki Y, Gillette S, Fong D, Ibrahim PN, Artis DR, Bollag G, Milburn MV, Kim SH, Schlessinger J, Zhang KY (2005) Nat Biotechnol 23:201–207
Lyne PD, Lamb ML, Saeh JC (2006) J Med Chem 49:4805–4808
Abel R, Young T, Farid R, Berne BJ, Friesner RA (2008) J Am Chem Soc 130:2817–2831
Beveridge DL, DiCapua FM, Beveridge DL, DiCapua FM (1989) Annu Rev Biophys Biophys Chem 18:431–492
Kollman P (1993) Chem Rev 93:2395–2417
Jarzynski C (1997) Phys Rev Lett 78:2690–2693
Shirts MR, Bair E, Hooker G, Pande VS (2003) Phys Rev Lett 91:140601
Fujitani H, Tanida Y, Matsuura A (2009) Phys Rev E 79:021914
Tanida Y, Ito M, Fujitani H (2007) Chem Phys 337:135–143
Hattori K, Kido Y, Yamamoto H, Ishida J, Kamijo K, Murano K, Ohkubo M, Kinoshita T, Iwashita A, Mihara K, Yamazaki S, Matsuoka N, Teramura Y, Miyake H (2004) J Med Chem 47:4151–4154
Kinoshita T, Nakanishi I, Warizuka M, Iwashita Y, Kido Y, Hattori K, Fujii T (2004) FEBS Lett 556:43–46
MOE is a product of Chemical Computing Group, Inc, 1010 Sherbrooke Street West, Suite 910, Montreal, Quebec, Canada
Gold is a product of a collaboration between the University of Sheffield, GlaxoSmithKline plc and Cambridge Structural Database System (CCDC). It is available from CCDC (12 Union Road, Cambridge, CB2 1EZ, UK)
Berendsen HJC, van der Spoel D, van Drunen R (1995) Comput Phys Commun 91:43–46
Lindahl E, Hess B, van der Spoel D (2001) J Mol Model 7:306–317
van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC (2005) J Comput Chem 26:1701–1718
Schmidt MW, Baldridge KK, Boatz JA, Elbert ST, Gordon MS, Jensen JH, Koseki S, Matsunaga N, Nguyen KA, Su S, Windus TL, Dupuis M, Montgomery JA (1993) J Comput Chem 14:1347–1363
Gordon MS, Schmidt MW (2005) Theory and Applications of Computational Chemistry: the first forty years. Elsevier, Amsterdam
Bayly CI, Cieplak P, Cornell W, Kollman PA (1993) J Phys Chem 97:10269–10280
Wang J, Wang W, Kollman PA, Case DA (2006) J Mol Graph Model 25:247–260
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) J Comput Chem 25:1157–1174
Fujitani H, Matsuura A, Sakai S, Sato H, Tanida Y (2009) J Chem Theor Comput 5:1155–1156
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) J Chem Phys 79:926–935
Nosé S (1984) Mol Phys 52:255–268
Hoover WG (1985) Phys Rev A 31:1695–1697
Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690
Hess B, Bekker H, Berendsen HJC, Fraaije JGEM (1997) J Comput Chem 18:1463–1472
Case DA, Darden TA, Cheatham TE III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, Walker RC, Zhang W, Wang B, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Schafmeister C, Ross WS, Kollman PA (2006) AMBER 9. University of California, San Francisco
Chang C, Chen W, Gilson MK (2005) J Chem Theor Comput 1:1017–1028
Acknowledgments
The authors thank E. Lindahl for help in modifying GROMACS. The authors thank Ms. Mariko Katsuyama, Mr. Makoto Takeuchi, Dr. Yuzo Matsumoto, Ms. Naoko Katayama, Dr. Makoto Oku, Ms. Ayako Moritomo, Dr. Kenichi Mori, and Dr. Hideyoshi Fuji for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Additional information
Kazuki Ohno and Takashi Mitsui equally contribute to this work.
Rights and permissions
About this article
Cite this article
Ohno, K., Mitsui, T., Tanida, Y. et al. Docking study and binding free energy calculation of poly (ADP-ribose) polymerase inhibitors. J Mol Model 17, 383–389 (2011). https://doi.org/10.1007/s00894-010-0728-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-010-0728-2