Abstract
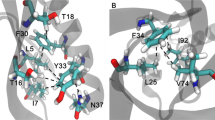
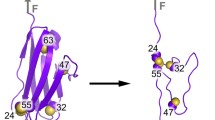
Mammalian metallothioneins (\( {\text{M}}_7^{\text{IIMTs}} \)) show a clustered arrangement of the metal ions and a nonregular protein structure. The solution structures of Cd3-thiolate cluster containing β-domain of mouse β-MT-1 and rat β-MT-2 show high structural similarities, but widely differing structure dynamics. Molecular dynamics simulations revealed a substantially increased number of \( {\text{NH - }}{{\text{S}}^\gamma } \) hydrogen bonds in β-MT-2, features likely responsible for the increased stability of the Cd3-thiolate cluster and the enfolding protein domain. Alterations in the \( {\text{NH - }}{{\text{S}}^\gamma } \) hydrogen-bonding network may provide a rationale for the differences in dynamic properties encountered in the β-domains of MT-1, -2, and -3 isoforms, believed to be essential for their different biological function.




Similar content being viewed by others
Abbreviations
- MT:
-
Metallothionein
- β-MT:
-
β-domain of metallothionein
- MD:
-
Molecular dynamics
- RMSD:
-
Root-mean-square deviation
- aa:
-
Amino acid
- H-bond:
-
Hydrogen bond
- NOE:
-
Nuclear Overhauser effect
References
Romero-Isart N, Vašák M (2002) Advances in the structure and chemistry of metallothioneins. J Inorg Biochem 88:388–396
Vašák M, Romero-Isart N (2005) In: King RB (ed) Encyclopedia of Inorganic Chemistry, 2nd. Wiley, New York, pp 3208–3221
Uchida Y, Takio K, Titani K, Ihara Y, Tomonaga M (1991) The growth inhibitory factor that is deficient in the Alzheimer’s disease brain is a 68 amino acid metallothionein-like protein. Neuron 7:337–347
Palmiter RD (1998) The elusive function of metallothioneins. Proc Natl Acad Sci USA 95:8428–8430
Miles A, Hawksworth G, Beattie J, Rodilla V (2000) Induction, regulation, degradation, and biological significance of mammalian metallothioneins. Crit Rev Biochem Mol Biol 35:35–70
Meloni G, Sonois V, Delaine T, Guilloreau L, Gillet A, Teissie J, Faller P, Vašák M (2008) Metal swap between Zn7-metallothionein-3 and amyloid-beta-Cu protects against amyloid-beta toxicity. Nat Chem Biol 4:366–372
Hidalgo J, Aschner M, Zatta P, Vašák M (2001) Roles of the metallothionein family of proteins in the central nervous system. Brain Res Bull 55:133–145
Schultze P, Wörgötter E, Braun W, Wagner G, Vašák M, Kägi JH, Wüthrich K (1988) Conformation of [Cd7]-metallothionein-2 from rat liver in aqueous solution determined by nuclear magnetic resonance spectroscopy. J Mol Biol 203:251–268
Zangger K, Öz G, Otvos JD, Armitage IM (1999) Three-dimensional solution structure of mouse [Cd7]-metallothionein-1 by homonuclear and heteronuclear NMR spectroscopy. Protein Sci 8:2630–2638
Robbins AH, McRee DE, Williamson M, Collett SA, Xuong NH, Furey WF, Wang BC, Stout CD (1991) Refined crystal structure of Cd, Zn metallothionein at 2.0 A resolution. J Mol Biol 221:1269–1293
Berweger CD, Thiel W, Van Gunsteren WF (2000) Molecular-dynamics simulation of the beta domain of metallothionein with a semi-empirical treatment of the metal core. Proteins 41:299–315
Chan J, Huang Z, Merrified M, Salgado M, Stillman M (2002) Studies of metal binding reactions in metallothioneins by spectroscopic, molecular biology, and molecular modeling techniques. Coord Chem Rev 233:319–339
Chan J, Merrifield ME, Soldatov AV, Stillman MJ (2005) XAFS spectral analysis of the cadmium coordination geometry in cadmium thiolate clusters in metallothionein. Inorg Chem 44:4923–4933
Öz G, Zangger K, Armitage IM (2001) Three-dimensional structure and dynamics of a brain specific growth inhibitory factor:metallothionein-3. Biochem 40:11433–11441
Wang H, Zhang Q, Cai B, Li H, Sze KH, Huang ZX, Wu HM, Sun H (2006) Solution structure and dynamics of human metallothionein-3 (MT-3). FEBS Lett 580:795–800
Ni FY, Cai B, Ding ZC, Zheng F, Zhang MJ, Wu HM, Sun HZ, Huang ZX (2007) Structural prediction of the beta-domain of metallothionein-3 by molecular dynamics simulation. Proteins 68:255–266
Romero-Isart N, Jensen LT, Zerbe O, Winge DR, Vašák M (2002) Engineering of metallothionein-3 neuroinhibitory activity into the inactive isoform metallothionein-1. J Biol Chem 277:37023–37028
Hasler DW, Jensen LT, Zerbe O, Winge DR, Vašák M (2000) Effect of the two conserved prolines of human growth inhibitory factor (metallothionein-3) on its biological activity and structure fluctuation: comparison with a mutant protein. Biochem 39:14567–14575
Soares TA, Daura X, Oostenbrink C, Smith LJ, Van Gunsteren WF (2004) Validation of the GROMOS force-field parameter set 45Alpha3 against nuclear magnetic resonance data of hen egg lysozyme. J Biomol NMR 30:407–422
Daura X, Oliva B, Querol E, Aviles FX, Tapia O (1996) On the sensitivity of MD trajectories to changes in water-protein interaction parameters:the potato carboxypeptidase inhibitor in water as a test case for the GROMOS force field. Proteins 25:89–103
Ulrich P, Scott W, Van Gunsteren WF, Torda AE (1997) Protein structure prediction force fields: parametrization with quasi-newtonian dynamics. Proteins 27:367–384
Sakharov DV, Lim C (2005) Zn protein simulations including charge transfer and local polarization effects. J Am Chem Soc 127:4921–4929
Sakharov DV, Lim C (2009) Force fields including charge transfer and local polarization effects: Application to proteins containing multi/heavy metal ions. J Comput Chem 30:191–202
Hünenberger P, Van Gunsteren WF (1998) Alternative schemes for the inclusion of a reaction-field correction into molecular dynamics simulations: Influence on the simulated energetic, structural, and dielectric properties of liquid water. J Chem Phys 108:6117–6134
Christen M, Van Gunsteren WF (2005) An approximate but fast method to impose flexible distance constraints in molecular dynamics simulations. J Chem Phys 122:144106
Van Gunsteren WF, Karplus M (1981) Effect of constraints, solvent and crystal environment on protein dynamics. Nature 293:677–678
Adman E, Watenpaugh KD, Jensen LH (1975) NH—-S hydrogen bonds in Peptococcus aerogenes ferredoxin, Clostridium pasteurianum rubredoxin, and Chromatium high potential iron protein. Proc Natl Acad Sci USA 72:4854–4858
Schäfer H, Smith LJ, Mark A, Van Gunsteren WF (2002) Entropy calculations on the molten globule state of a protein: side-chain entropies of a-Lactoalbumin. Proteins 46:215–224
Nolde S, Arseniev A, Orekhov VY, Billeter M (2002) Essential domain motions in barnase revealed by MD simulations. Proteins 46:250–258
Gargallo R, Hünenberger P, Avilés F, Oliva B, Sci P (2003) Molecular dynamics simulation of highly charged proteins: comparison of the particle-particle particle-mesh and reaction field methods for the calculation of electrostatic interactions. Protein Sci 10:2161–2172
Peter C, Oostenbrink C, Van Dorp A, Van Gunsteren WF (2004) Estimating entropies from molecular dynamics simulations. J Chem Phys 120:2652–2661
Rayment I, Wesenberg G, Meyer TE, Cusanovich MA, Holden HM (1992) Three-dimensional structure of the high-potential iron-sulfur protein isolated from the purple phototrophic bacterium Rhodocyclus tenuis determined and refined at 1.5 A resolution. J Mol Biol 228:672–686
Marmorstein R, Carey M, Ptashne M, Harrison SC (1992) DNA recognition by GAL4: structure of a protein-DNA complex. Nature 356:408–414
Blake PR, Summers MF (1994) Insights into the structural and electronic properties of metalloproteins by heteronuclear magnetic resonance. Adv Inorg Biochem 10:201–228
Berg J (1995) Zinc Finger Domains: From Predictions to Design. Acc Chem Res 28:14–19
Jensen L (1977) Crystal and Molecular Structure of Rubredoxin from Clostridium pasteurianum. In W Lovenberg (ed) Iron-sulfur proteins Academic Press, New York, p 184
Smith JN, Hoffman JT, Shirin Z, Carrano CJ (2005) H-bonding interactions and control of thiolate nucleophilicity and specificity in model complexes of zinc metalloproteins. Inorg Chem 44:2012–2017
Maynard AT, Covell DG (2001) Reactivity of zinc finger cores: analysis of protein packing and electrostatic screening. J Am Chem Soc 123:1047–1058
Acknowledgments
This work was supported by the Swiss National Science Foundation Grant 3100A0-111884 (to M.V), the Spanish Ministerio de Educación y Ciencia (PROFIT PSE0100000-2007-1) and Spanish Ministerio de Ciencia e Innovación (BIO2008-0205) (to B.O), and Fundación Ramón Areces, Spain, postdoctoral fellowship (to N.R.I).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary materials
Below is the link to the electronic supplementary material.
Fig. S1
Experimental structures and averaged structures after the simulations of β-MT-1 and β-MT-2 are shown. \( {{\text{C}}^\alpha } \) trace of the averaged conformations (in white) of Cd3β-MT-1 (a) and Cd3β-MT-2 (b) obtained from the last 4 ns of MD simulation superimposed with their respective experimental NMR structures (cyan). Metal-thiolate clusters resulting from the superimpositions are shown as balls and sticks. (DOC 6212 kb)
Fig. S2
Time series of the electrostatic energy components of (a) Cd3β-MT-1 and (b) Cd3β-MT-2 during the 5 ns MD simulations. Energetic components are grouped according to the chemical origin of the interaction: protein refers to atoms of the amino acid residues and cadmium refers to Cd2+ atoms. Continuous line: protein-protein and protein-cadmium interactions; line plus circles: cadmium-cadmium interactions. (DOC 3747 kb)
Fig. S3
Time series (ps) of RMS deviations (Å) of the backbone (black) and all (red) atoms of the S6P,S8P+T5 mutant of Cd3β-MT-1. All combinations of isomeric states of prolyl residues are considered: a) trans-trans; b) trans-cis; c) cis-trans; and d) cis-cis. (DOC 3343 kb)
Fig. S4
\( {{\text{C}}_\alpha } \) atom-positional RMS fluctuations (Å) per residue around the averaged structures in the last 4ns of the S6P,S8P+T5 mutant of Cd3β-MT-1. All combinations of isomeric states of prolyl residues are considered: a) trans-trans; b) trans-cis; c) cis-trans; and d) cis-cis. (DOC 1180 kb)
Table S1
Hydrogen bonds present in the S6P,S8P+T5 mutant of Cd3β-MT-1 with percentages larger than 50% of hydrogen bond formation between donor and acceptor atoms along the equilibrium time of the simulation (4 ns). All combinations of isomeric states of prolyl residues are considered: trans-trans, trans-cis, cis-trans and cis-cis. \( {\text{NH - }}{{\text{S}}^\gamma } \) H-bonds are highlighted in bold. (DOC 42 kb)
Table S2
Metal-thiolate cluster topology of Cd3Cys9 in the S6P,S8P+T5 mutant of Cd3β-MT-1. All combinations of isomeric states of prolyl residues are considered: transtrans, trans-cis, cis-trans and cis-cis. Time-averaged and fluctuations (SD) of bond lengths and bond angles between bridging (Sb) or terminal (St) sulfur atoms and cadmium ions (Cd). The calculated virtual entropy shows the local disorder of the cluster atoms Cd3S9. (DOC 39 kb)
Rights and permissions
About this article
Cite this article
Romero-Isart, N., Oliva, B. & Vašák, M. Influence of \( {\text{NH - }}{{\text{S}}^\gamma } \) bonding interactions on the structure and dynamics of metallothioneins. J Mol Model 16, 387–394 (2010). https://doi.org/10.1007/s00894-009-0542-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-009-0542-x