Abstract
Glucagon-like peptide-1 receptor (GLP-1R) is a promising molecular target for developing drugs treating type 2 diabetes. We have predicted the complete three-dimensional structure of GLP-1R and the binding modes of several GLP-1R agonists, including GLP-1, Boc5, and Cpd1, through a combination of homology modeling, molecular docking, and long-time molecular dynamics simulation on a lipid bilayer. Our model can reasonably interpret the results of a number of mutation experiments regarding GLP-1R as well as the successful modification to GLP-1 by Liraglutide. Our model is also validated by a recently revealed crystal structure of the extracellular domain of GLP-1R. An activation mechanism of GLP-1R agonists is proposed based on the principal component analysis and normal mode analysis on our predicted GLP-1R structure. Before the complete structure of GLP-1R is determined through experimental means, our model may serve as a valuable reference for characterizing the interactions between GLP-1R and its agonists.

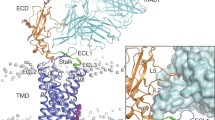
Comparison of our predicted model of rGLP-1R (left) with the recently revealed crystal structure of hGLP-1R (right)














Similar content being viewed by others
References
Murphy KG, Bloom SR (2007) Proc Natl Acad Sci USA 104:689–690. doi:10.1073/pnas.0610679104
Drucker DJ (2007) Endocrinology 142:521–527. doi:10.1210/en.142.2.521
Demuth HU, McIntosh CH, Pederson RA (2005) Biochim Biophys Acta 1751:33–44
Madsbad S, Schmitz O, Ranstam J, Jakobsen G, Matthews DR (2004) Diabetes Care 27:1335–1342. doi:10.2337/diacare.27.6.1335
Chen DS, Liao JY, Li N, Zhou CH, Liu Q, Wang MW et al. (2007) Proc Natl Acad Sci USA 104:943–948. doi:10.1073/pnas.0610173104
Knudsen LB, Kiel D, Teng M, Behrens C, Bhumralkar D, Kodra JT et al. (2007) Proc Natl Acad Sci USA 104:937–942. doi:10.1073/pnas.0605701104
Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT (2005) Nucleic Acids Res 33:36–38. doi:10.1093/nar/gki410
Xiao Q, Jeng W, Wheeler MB (2000) J Mol Endoc 25:321–335. doi:10.1677/jme.0.0250321
Philip Bourne TNB, Feng ZK, Gilliland G, Jain S, Ravichandran V, Schneider B et al. (2001) Nucleic Acids Res 29:214–218. doi:10.1093/nar/29.1.214
Pearson WR (1990) Methods Enzymol 183:63–98. doi:10.1016/0076-6879(90)83007-V
Marti-Renom MA, Stuart A, Fiser A, Sánchez R, Melo F, Sali A (2000) Annu Rev Biophys Biomol Struct 29:291–325. doi:10.1146/annurev.biophys.29.1.291
Discovery Studio software (version 2.0) Accelyrs Inc. San Diego CA U.S.A. (2007)
Bazarsuren A, Grauschopf U, Wozny M, Reusch D, Hoffmann E, Schaefer W et al. (2002) Biophys Chem 96:305–318. doi:10.1016/S0301-4622(02)00023-6
Xiao Q, Jeng W, Wheeler MB (2000) J Mol Endoc. 25:321–335. doi:10.1677/jme.0.0250321
Suleiman AS, Donnelly D (2003) FEBS Lett 553:342–346. doi:10.1016/S0014-5793(03)01043-3
Maturana RL, Donnelly D (2002) FEBS Lett 530:244–248. doi:10.1016/S0014-5793(02)03492-0
Runge S, Gram C, Hans BO, Madsen K, Knudsen LB, Wulff BS (2003) J Biol Chem 278:28005–28010. doi:10.1074/jbc.M301085200
Pan CQ, Buxton JM, Yung SL, Tom I, Yang L, Chen HX et al. (2006) J Biol Chem 281:12506–12515. doi:10.1074/jbc.M600127200
Adelhorst K, Hedegaard BB, Knudsen LB, Kirks O (1994) J Biol Chem 269:6276–6278
Gallwitz B, Witt M, Paetzold G, Wortmann CM, Zimmermann B, Eckart K et al (1994) Eur J Biochem 225:1151–1156. doi:10.1111/j.1432-1033.1994.1151b.x
Wilmen A, Eyll BV, Goke B, Goke R (1997) Peptides 18:301–305. doi:10.1016/S0196-9781(96)00321-X
Jones DT, Taylor WR, Thornton JM (1994) Biochemistry 33:3038–3049. doi:10.1021/bi00176a037
Moller S, Croning MDR, Apweiler R (2001) Bioinformatics 17:646–653. doi:10.1093/bioinformatics/17.7.646
Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A (1997) Protein Eng 10:673–676. doi:10.1093/protein/10.6.673
Schultz J, Milpetz F, Bork P, Ponting CP (1998) Proc Natl Acad Sci USA 95:5857–5864. doi:10.1073/pnas.95.11.5857
Arai M, Mitsuke H, Ikeda M, Xia JX, Kikuchi T, Satake M et al. (2004) Nucleic Acids Res 32:390–393. doi:10.1093/nar/gkh380
Tusnady GE, Simon I (1998) J Mol Biol 283:489–506. doi:10.1006/jmbi.1998.2107
von Heijne G (1992) J Mol Biol 225:487–494. doi:10.1016/0022-2836(92)90934-C
Bagos PG, Liakopoulos TD, Hamodrakas SJ (2006) BMC Bioinform 7:189–206. doi:10.1186/1471-2105-7-189
Hirokawa T, Boon-Chieng S, Mitaku S (1998) Bioinformatics 14:378–379. doi:10.1093/bioinformatics/14.4.378
Persson B, Argos P (1997) J Protein Chem 16:453–457. doi:10.1023/A:1026353225758
Pashou EE, Litou ZI, Liakopoulos TD, Hamodrakas SJ (2004) In Silico Biol 4:0012–0012
Taylor PD, Attwood TK, Flower DR (2003) Nucleic Acids Res 31:3698–3700. doi:10.1093/nar/gkg554
Juretic D, Zoranic L, Zucic D (2002) J Chem Inf Comput Sci 42:620–632. doi:10.1021/ci010263s
Unson CG (2002) Biopolymers 66:218–235. doi:10.1002/bip.10259
Maturana RL, Donnelly D (2002) FEBS Lett 530:244–248. doi:10.1016/S0014-5793(02)03492-0
Runge S, Gram C, Brauner-Osborne H, Madsen K, Knudsen LB, Wulff BS (2003) J Biol Chem 278:28005–28010. doi:10.1074/jbc.M301085200
Maturana RL, Willshaw A, Kuntzsch A, Rudolph R, Donnelly D (2003) J Biol Chem 278:10195–10200. doi:10.1074/jbc.M212147200
Suleiman AS, Donnelly D (2003) Br J Pharmacol 140:339–346. doi:10.1038/sj.bjp.0705453
Gallwitz B, Witt M, Paetzold G, Wortmann CM, Zimmermann B, Eckart K et al. (1994) Eur J Biochem 225:1151–1156. doi:10.1111/j.1432-1033.1994.1151b.x
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) J Appl Cryst 26:283–291. doi:10.1107/S0021889892009944
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR et al. farkas O, Tomasi J, Barone V, Cossi M, Cammi R, Mennucci B, Pomelli C, Adamo C, Clifford S, Ochterski J, Petersson GA, Ayala PY, Cui Q, Morokuma K, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Cioslowski J, V Rotiz J, Stefanov BB, Liu G, Liashenko A, Piskora P, Komaromi I, Gomperts R, Martin RL, Fox DJ, Keith T, AlLaham MA, Peng CY, Nanayakkara A, Gonzalez C, Challacombe M, Gill MW, Johnson B, Chen W, Wong MW, Andres JL, Gonzalez C, Gordon MH, Replogle ES, Pople JA Gaussian, Inc., Pittsburgh PA (2003)
The SYBYL software (version 7.2) Tripos Inc., St. Louis, Missouri (2006)
Jones G, Willett P, Glen RC, Leach AR, Taylor R (1997) J Mol Biol 267:727–748. doi:10.1006/jmbi.1996.0897
van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC (2005) J Comput Chem 26:1701–1718. doi:10.1002/jcc.20291
Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR (1984) J Chem Phys 81:3684–3690. doi:10.1063/1.448118
Berger O, Edholm O, Jähnig F (1997) Biophys J 72:2002–2013
Darden T, York D, Pedersen L (1993) J Chem Phys 98:10089–10092. doi:10.1063/1.464397
Ryckaert JP, Ciccotti G, Berendsen HJC (1977) J Comput Phys 23:327–341. doi:10.1016/0021-9991(77)90098-5
Suhre K, Sanejouand YH (2004) Nucleic Acids Res 32:610–614. doi:10.1093/nar/gkh368
Runge S, Thogersen H, Madsen K, Lau J, Rudolph R (2008) J Biol Chem 283:11340–11347. doi:10.1074/jbc.M708740200
Acknowledgment
The authors are grateful to the financial supports from the Chinese National Natural Science Foundation (grants 20502031 & 20772149), the Chinese Ministry of Science and Technology (grant 2006AA02Z337), and the Science and Technology Commission of Shanghai Municipality (grants 06PJ14115 & 074319113).
Supporting information available
Three-dimensional structures of the GLP-1R models described in this manuscript are available from the authors upon request.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lin, F., Wang, R. Molecular modeling of the three-dimensional structure of GLP-1R and its interactions with several agonists. J Mol Model 15, 53–65 (2009). https://doi.org/10.1007/s00894-008-0372-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-008-0372-2