Abstract
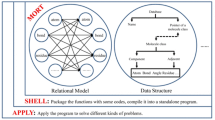
Structural characterization of enzymes that belong to microbial metabolic pathways is very important for structure-based drug design since some of these proteins may be present in the bacterial genome, but absent in humans. Thus, metabolic pathways became potential targets for drug design. The motivation of this work is the fact that Mycobacterium tuberculosis is the cause of the deaths of millions of people in the world, so that the structural characterization of protein targets to propose new drugs has become essential. DBMODELING is a relational database, created to highlight the importance of methods of molecular modeling applied to the Mycobacterium tuberculosis genome with the aim of proposing protein-ligand docking analysis. There are currently more than 300 models for proteins from Mycobacterium tuberculosis genome in the database. The database contains a detailed description of the reaction catalyzed by each enzyme and their atomic coordinates. Information about structures, a tool for animated gif image, a table with a specification of the metabolic pathway, modeled protein, inputs used in modeling, and analysis methods used in this project are available in the database for download. The search tool can be used for reseachers to find specific pathways or enzymes.
Figure The shikimate pathway in the sequence of seven metabolic steps, from phosphoenolpyruvate and erythrose-4-phosphate until the conversion to chorismate.





Similar content being viewed by others
References
Snider DE, Raviglione M, Kochi A (1994) In: Bloom BR (ed) Tuberculosis: pathogenesis, protection, and control, Vol 2(11). Am Soc Microbiol, Washington
Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry III CE, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell BG (1998) Nature 393:537–544
McKinney DJ, Jacobs Jr WR, Bloom BR (1998) Persisting problems in tuberculosis. In: Krause RM (ed) Emerging infections. Academic, New York, pp 51–146
Campos HS (1999) Boletim de Pneumologia Sanitária 7:51–64
Bloom BR, Murray CJL (1992) Science 257:1055–1064
Basso LA, Blanchard JS (1998) Adv Exp Med Biol 456:115–144
de Azevedo Jr WF, Mueller-Dieckmann HJ, Schulze-Gahmen U, Worland PJ, Sausville E, Kim SH (1996) Proc Natl Acad Sci USA 93:2735–2740
de Azevedo Jr WF, Leclerc S, Meijer L, Havlicek L, Strnad M, Kim SH (1997) Eur J Biochem 243:518–526
Foster MJ (2002) Micron 33:365–384
Karp PD (2001) Science 293:2040–2044
Browne WJ, North AC, Phillips DC, Drew K, Vanaman TC, Hill RL (1969) J Mol Biol 42:65–86
Sali A, Blundell TL (1993) J Mol Biol 234:779–815
Sánchez R, Sali A (1999) Bioinformatics 15:1060–1061
Gerstein M, Levitt M (1997) Proc Nat Acad Sci USA 94:11911–11916
Fischer D, Eisenberg D (1999) Curr Opin Struct Biol 9:208–211
Uchôa HB, Jorge GE, da Silveira NJF, Camera Jr JC, de Azevedo Jr WF (2004) Biochem Biophys Res Commun 325:1481–1486
Laskowski RA, MacArthur MW, Thornton JM (1998) Curr Opin Struct Biol 8:631–639
Wilson C, Gregoret LM, Agard DA (1993) J Mol Biol 229:996–1006
Laskowski RA, MacArthurm MW, SmithDK, Jones DT, Hutchinson EG, Morris AL, Naylor D, Moss DS, Thornton JM (1994) PROCHECK v.3.0—Program to check the stereochemistry quality of protein structures—Operating instructions
Hooft RW, Sander C, Vriend G (1996a) Proteins 26:363–376
Bowie JU, Luthy R, Eisenberg D (1991) Science 253:164–170
Kabsch W, Sander C (1983) Biopolymers 22:2577–2637
Sippl MJ (1990) J Mol Biol 213:859–883
Luthy R, Bowie JU, Eisenberg K (1992) Nature 356:83–85
Arcuri HA, Canduri F, Pereira JH, da Silveira NJF, Camera Jr JC, de Oliveira JS, Basso LA, Palma MS, Santos DS, de Azevedo Jr WF (2004) Biochem Biophys Res Commun 320:979–991
da Silveira NJF, Uchôa HB, Canduri F, Pereira JH, Camera Jr JC, Basso LA, Palma MS, Santos DS, de Azevedo Jr WF (2004) Biochem Biophys Res Commun 322:100–104
Brünger AT (1992) X-PLOR, a System for crystallography and NMR, Version 3.1.Yale University Press, New Haven
EU 3-D Validation Network (1998) J Mol Biol 276:417–436
DuBois P (2000) MySQL. New Riders, Indianapolis
Wall L, Christiansen T, Orwant J (2000) In: O’Reilly, Associates Inc (eds) Programming Perl, 3rd edn
Pieper U, Eswar N, Braberg H, Madhusudhan MS, Davis FP, Stuart AC, Mirkovic N, Rossi A, Marti-Renom MA, Fiser A, Webb B, Greenblatt D, Huang CC, Ferrin TE, Sali A (2004) Nucleic Acids Res 32:D217-D222
Kawabata T, Fukuchi S, Homma K, Ota M, Araki J, Ito T, Ichiyoshi N, Nishikawa K (2002) Nucleic Acids Res 30:294–298
Bairoch A, Apweiler R (1999) Nucleic Acids Res 27:49–54
Westbrook J, Feng Z, Chen L, Yang H, Berman HM (2003) Nucleic Acids Res 31:489–491
Abola BB, Bernstein FC, Bryant SH, Koetzle T, Weng J (1987) Protein data bank. In: Allen FH, Bergerhoff G, Sievers R (eds) Crystallographic databases-information, content, software systems, scientific applications. Data Commission of the International Union of Crystallography, Bonn Cambridge Chester, pp 107–132
Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M (1999) Nucleic Acids Res 27:29–34
Karp PD, Riley M, Paley SM, Pellegrini-Toole A (2002) Nucleic Acids Res 30:59–61
Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A (2000) Annu Rev Biophys Biomol Struct 29:291–325
Sanchez R, Sali A (1998) Proc Natl Acad Sci USA 95:13597–13602
Koehl P, Levitt M (1999) Nature Struct Biol 6:108–111
Coggins JR, Abell C, Evans LB, Frederickson M, Robinson DA, Roszak AW, Lapthorn AP (2003) Biochem Soc Trans 31:548–552
Campbell SA, Richards TA, Mui EJ, Samuel BV, Coggins JR, McLeod R, Roberts CW (2004) Int J Parasitol 34:5–13
Herman KM, Weaver LM (1999) Annu Rev Plant Mol Biol 50:473–503
de Azevedo Jr WF, Canduri F, de Oliveira JS, Basso LA, Palma MS, Pereira JH, Santos DS (2002) Biochem Biophys Res Commun 295:142–148
Pereira JH, Canduri F, de Oliveira JS, da Silveira NJF, Basso LA, Palma MS, de Azevedo Jr WF, Santos DS (2003) Biochem Biophys Res Commun 312:608–614
Pereira JH, Oliveira JS, Canduri F, Dias MVB, Palma MS, Basso LA, de Azevedo Jr WF, Santos DS (2004) Biochem Biophys Res Commun 325:10–17
Pereira JH, Oliveira JS, Canduri F, Dias MVB, Palma MS, Basso LA, Santos DS, de Azevedo Jr WF (2004) Acta Crystallogr Sect D-Biol Crystallogr 60:2310–2319
Chakrabarti S, John J, Sowdhamini R (2004) J Mol Model 10:69–75
Acknowledgements
This work was supported by grants from FAPESP (Process Numbers: 02/10239–6, SMOLBNet 01/07532–0, 02/04383–7, 04/00217–0), CNPq, CAPES and Instituto do Milênio (CNPq-MCT). WFA, LAB, MSP, and DSS are researchers for the Brazilian Council for Scientific and Technological Development.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Silveira, N.J.F.d., Uchôa, H.B., Pereira, J.H. et al. Molecular models of protein targets from Mycobacterium tuberculosis. J Mol Model 11, 160–166 (2005). https://doi.org/10.1007/s00894-005-0240-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-005-0240-2